Predicting Cervical Intraepithelial Neoplasia Grades Using Clinical, Virological, and Immunocytochemical Data: A Retrospective Study from Eastern Europe
by Iulian-Valentin Munteanu1, Demetra Socolov2, Razvan Socolov2, Ana-Maria Adam1, Gigi Adam3, Ingrid-Andrada Vasilache2*, Petronela Vicoveanu4, Valeriu Harabor1, Anamaria Harabor1, and Alina-Mihaela Calin1
1Clinical and Surgical Department, Faculty of Medicine and Pharmacy, ‘Dunarea de Jos’ University, 800216 Galati, Romania
2Department of Mother and Child Care “Grigore T. Popa” University of Medicine and Pharmacy Iasi, Romania
3Department of Pharmaceutical Sciences, Faculty of Medicine and Pharmacy, ‘Dunarea de Jos’ University, 800216 Galati, Romania
4Department of Mother and Newborn Care, Faculty of Medicine and Biological Sciences, ‘Ștefan cel Mare’ University, 720229, Suceava, Romania
*Corresponding Author: Ingrid-Andrada Vasilache, Department of Mother and Child Care “Grigore T. Popa” University of Medicine and Pharmacy Iasi, Romania.
Received Date: 30 July 2025
Accepted Date: 04 August 2025
Published Date: 06 August 2025
Citation: Munteanu IV, Socolov D, Socolov R, Adam AM, Adam G, et al. (2025) Predicting Cervical Intraepithelial Neoplasia Grades Using Clinical, Virological, and Immunocytochemical Data: A Retrospective Study from Eastern Europe. Gynecol Obstet Open Acc 9: 242. https://doi.org/10.29011/2577-2236.100242.
Abstract
While histopathological examination remains the gold standard, integration of clinical, cytological, and virological parameters may enable non-invasive prediction of cervical intraepithelial neoplasia (CIN) through machine learning (ML) approaches. The aim of this study was to evaluate the performance of multiple machine learning algorithms in predicting CIN1, CIN2, and CIN3 using routinely collected clinical and laboratory data. This retrospective study included 98 women who underwent colposcopy-guided cervical biopsies at a tertiary care center in Romania between January 2022 and December 2024. Features analyzed included age, smoking, sexual behaviour, HPV genotype, cytological findings, and CINtec® (p16/Ki-67) dual staining status. Data were balanced using SMOTE, and models were trained using five-fold cross-validation. Predictive performance was assessed using accuracy, precision, recall, F1-score, and the area under the receiver operating characteristic curve (ROC AUC) for each ML model. Among 98 women, 53 (54.1%) had CIN1, 36 (36.7%) CIN2, and 9 (9.2%) CIN3. Random Forest achieved the best performance in predicting CIN1 (accuracy 63%, F1-score 0.67, ROC AUC 0.658). Logistic Regression with L2 regularization outperformed other models in predicting CIN2 (accuracy 89%, F1-score 0.77, ROC AUC 0.839). For CIN3 prediction, Logistic Regression again showed the highest performance (accuracy 96%, F1-score 0.80, ROC AUC 0.98). CatBoost and XGBoost showed competitive performance in predicting CIN2 and CIN3, while Naive Bayes and SVM exhibited variable performance depending on the CIN grade. Machine learning models, particularly Logistic Regression and ensemble-based classifiers, demonstrated promising performance in predicting CIN grades using readily available clinical and laboratory data.
Keywords: cervical dysplasia; cytologic dual-stain; machine learning algorithms; diagnosis
Introduction
Cervical intraepithelial neoplasia (CIN) represents a spectrum of premalignant changes, and stratification into CIN1, CIN2, and CIN3 informs both surveillance and treatment strategies. However, current diagnostic pathways heavily rely on colposcopy and histopathology, which are invasive and resource-intensive. In Eastern Europe, the prevalence of HPV infection is estimated at 21%, with the region showing the highest global rates of highgrade cervical dysplasia (~4.3% overall in Europe) [1-3]. Among women with normal cytology in Central and Eastern Europe, HPV prevalence remains substantial at 12.6%, with HPV16 being the most frequently detected genotype, particularly in high-grade lesions [3].
Interobserver variability remains a major challenge in the histopathological diagnosis of cervical intraepithelial neoplasia (CIN), particularly in the evaluation of lower-grade lesions. Substantial disagreement among pathologists has been welldocumented, especially in differentiating between CIN I and CIN II. In contrast, diagnostic agreement tends to be higher for invasive carcinoma and CIN III. This is quantitatively reflected in reported kappa (κ) values, which measure interrater reliability: κ = 0.496 for CIN III, compared with κ = 0.172 and κ = 0.175 for CIN II and CIN I, respectively [4-6]. A key contributor to this variability is the morphological overlap between reactive epithelial changes and low-grade neoplastic processes, particularly CIN I. Histological features can often mimic one another, and even among experienced observers, the distinction may be ambiguous. As a result, both overdiagnosis and underdiagnosis of CIN I are common, which has implications for both patient anxiety and resource utilization [4-6].
To address these challenges, several emerging technologies have shown promise. Biomarker-based profiling—particularly the use of immunocytochemical markers such as p16, Ki-67, and DNA methylation signatures—has demonstrated the ability to enhance risk stratification and improve the specificity of CIN grading [7, 8]. These tools offer the potential to reduce overtreatment by identifying lesions with low malignant potential. In parallel, advances in artificial intelligence have introduced new diagnostic approaches. Deep learning models have achieved diagnostic accuracies as high as 90.8%, offering a reproducible and scalable method to support pathologist decision-making [9].
Recent advancements in machine learning (ML) and deep learning have markedly improved the diagnostic performance for cervical intraepithelial neoplasia, surpassing traditional approaches in many contexts. Diagnostic accuracy is influenced by multiple factors, including the nature of the input data, model architecture, and the robustness of the validation technique employed.
Ensemble convolutional neural networks (CNNs) trained on digital histopathological images have achieved exceptionally high accuracy, with reported rates up to 94.6% for CIN grading, underscoring their potential as decision-support tools in pathology workflows [10, 11].
Multimodal models combining clinical variables with colposcopic imaging data have further enhanced prediction capabilities. For instance, a convolutional neural network, which integrated both visual and clinical information, achieved a diagnostic accuracy of 92.3%, highlighting the value of contextual clinical data in improving classification outcomes [12]. In the context of highgrade squamous intraepithelial lesions (HSIL), the Swin-B model reached an accuracy of 91.4%, supporting its use in triaging potentially severe lesions [13].
Beyond imaging, ML models trained on molecular data have also shown strong discriminatory power. A Random Forest algorithm applied to DNA methylation profiles for identifying CIN2+ lesions achieved an area under the ROC curve (AUC) of 0.90, outperforming traditional methods such as HPV genotyping and cytological screening [14]. Similarly, a Naive Bayes classifier using methylation data demonstrated an AUC of 0.88 and a specificity of 93.9%, reflecting its utility in high-specificity applications [14]. Moreover, a recent meta-analysis of 77 studies evaluating artificial intelligence–assisted cytology reported pooled diagnostic accuracies ranging from 90% to 94%, further confirming the consistency and reliability of AI-enhanced diagnostic systems in cervical cancer screening programs [15].
The present study aimed to evaluate the predictive performance of various supervised machine learning models in differentiating between CIN1, CIN2, and CIN3 lesions, utilizing a dataset comprising HPV genotyping, cytology findings, and p16/Ki-67 dual staining.
Materials and methods
Sampling
Study Population
This retrospective study included 98 women who underwent colposcopy-guided cervical biopsies at tertiary care centers from Romania between January 2022 and December 2024. Patients were eligible for inclusion if they were adult women (aged ≥18 years) who underwent colposcopic evaluation and had histopathological confirmation of cervical intraepithelial neoplasia (CIN1, CIN2, or CIN3). Inclusion required availability of complete clinical data, including HPV genotyping results, cytological findings, and immunocytochemical staining (CINtec® for p16/Ki-67). Patients were excluded if they had a history of cervical cancer, previous hysterectomy, immunosuppressive disorders, or incomplete clinical or histopathologic data. Additionally, pregnant women and individuals with co-existing gynecologic malignancies were excluded to reduce confounding. The final dataset comprised 98 patients who met all eligibility criteria and were included in the analysis. Patients were classified into three histologically confirmed groups: CIN1 (n = 53), CIN2 (n = 36), and CIN3 (n = 9). Demographic and clinical variables, including smoking status, sexual history, HPV infection, cytological findings, and immunocytochemical markers, were collected from their medical records.
Feature Selection and Data Pre-processing
Clinical and laboratory variables with potential relevance to cervical neoplasia were extracted for analysis. These included age, residence (urban or rural), smoking history, number of sexual partners, age at sexual debut, parity, combined oral contraceptive (COC) use, sexually transmitted disease (STD) history, prior cervical treatment, HPV genotype (16/18, other high-risk, or lowrisk), CINtec® dual-staining status (p16/Ki-67), and Pap smear results (ASCUS, LSIL, HSIL, NILM). Continuous variables such as age were normalized, while categorical variables were onehot encoded. Missing data were imputed using median values for continuous variables and the most frequent category for categorical variables.
Machine Learning Models
To evaluate the predictive capacity of clinical and laboratory features in classifying CIN grade, supervised machine learning models were developed separately for each CIN category (CIN1, CIN2, CIN3). The following classifiers were employed: Logistic
Regression, Random Forest, Support Vector Machine (SVM),
Naive Bayes, k-Nearest Neighbors (KNN), Extreme Gradient Boosting (XGBoost), and CatBoost. Model development was performed using the scikit-learn and xgboost libraries in Python (version 3.11).
Handling Class Imbalance
Given the class imbalance—especially the low number of CIN3 cases—Synthetic Minority Oversampling Technique (SMOTE) was applied to the training data within a stratified k-fold crossvalidation framework (k = 5). The oversampling was applied only on the training set in each fold to avoid data leakage.
Hyperparameter Tuning
Hyperparameters for each model were optimized using grid search with cross-validation. For logistic regression, L2 and L1 regularization parameters (C values) were tuned. For random forest, the number of estimators and tree depth were varied. SVM models were optimized for the penalty term (C) and kernel type (linear or radial basis function). KNN models were tuned by varying the number of neighbors (k). Gradient boosting models (XGBoost and CatBoost) were optimized for learning rate, depth, number of estimators, and iterations as appropriate.
Evaluation Metrics
Model performance was evaluated on a hold-out test set, with metrics including precision, recall, F1-score, overall accuracy, and area under the receiver operating characteristic curve (ROC AUC). Metrics were calculated for each class (0 and 1), with particular focus on the performance for the positive class (presence of the respective CIN grade). Macro and weighted averages were also computed. The best-performing parameters and metrics for each classifier were recorded and are presented alongside visual representations.
Ethical Approval
This study was conducted in accordance with the principles of the Declaration of Helsinki. Institutional Review Board (IRB) approval was obtained from the Institutional Ethics Committee of Clinical Hospital of Obstetrics and Gynecology ,,Buna vestire” Galati (No. 115/05.01.2021). No financial incentives were provided for participation, and patient care was not influenced by study involvement.
Results
Patient characteristics
Ninety eight patients were included in the study and their clinical characteristics are presented in Table 1. The presence of multiple sexual partners showed a statistically significant association with higher CIN grades (p = 0.022). Only 1 patient (1.9%) in the CIN1 group reported multiple partners, whereas 6 (16.7%) in CIN2 and 2 (22.2%) in CIN3 reported the same behaviour. Early sexual debut (defined as ≤15 years) was more frequently reported in CIN2 and CIN3 groups, though the difference was not statistically significant (p = 0.165).
A significant association was observed between sexually transmitted disease history and CIN grade (p < 0.001). No patients in the CIN1 group had a history of STD, whereas 1 (2.8%) in CIN2 and 3 (33.3%) in CIN3 did.
HPV 16/18 infection was significantly more common in highergrade lesions (p < 0.001). Twelve patients (22.6%) with CIN1 had HPV 16/18, compared with 18 (50.0%) in CIN2 and 8 (88.9%) in CIN3. Low-risk HPV types were not significantly associated with CIN severity (p = 0.367), found in 10 (19.2%), 3 (8.6%), and 1 (11.1%) patients with CIN1, CIN2, and CIN3, respectively.
CINtec+ (p16/Ki-67 dual staining) positivity was strongly associated with higher CIN grade (p < 0.001). It was present in 6 (11.32%) of CIN1 cases, 6 (16.7%) of CIN2, and 5 (55.6%) of CIN3. Low-grade squamous intraepithelial lesion (LSIL) findings were more common in higher CIN grades (p = 0.034), reported in 24 (45.3%) CIN1 cases, 22 (61.1%) CIN2 cases, and 8 (88.9%) CIN3 cases. High-grade squamous intraepithelial lesion (HSIL)
results were significantly more common in CIN2 and CIN3 groups compared to CIN1 (p < 0.001). Only 2 patients (3.8%) in CIN1 had HSIL, while 16 (44.4%) in CIN2 and 5 (55.6%) in CIN3 did.
|
Variable |
CIN1 (n=53 patients) |
CIN2 (n=36 patients) |
CIN3 (n=9 patients) |
p-value |
|
Residence (Rural) |
22 (41.5%) |
15 (41.7%) |
3 (33.3%) |
0.891 |
|
Smoking (Yes) |
14 (26.4%) |
9 (25.0%) |
1 (11.1%) |
0.612 |
|
Multiple sexual partners |
1 (1.9%) |
6 (16.7%) |
2 (22.2%) |
0.022 |
|
Early Sexual Debut |
1 (1.9%) |
4 (11.1%) |
1 (11.1%) |
0.165 |
|
Multiparity |
21 (39.6%) |
14 (38.9%) |
6 (66.7%) |
0.779 |
|
COC Use (Yes) |
5 (9.4%) |
8 (22.2%) |
2 (22.2%) |
0.215 |
|
STD (Yes) |
0 (0.0%) |
1 (2.8%) |
3 (33.3%) |
<0.0001 |
|
Previous cervical treatment |
38 (79.2%) |
22 (61.1%) |
5 (55.6%) |
0.125 |
|
HPV 16/18 |
12 (22.6%) |
18 (50.0%) |
8 (88.9%) |
<0.0001 |
|
Other HR HPV |
23 (43.4%) |
22 (61.1%) |
2 (22.2%) |
0.070 |
|
Low-risk HPV |
10 (19.2%) |
3 (8.6%) |
1 (11.1%) |
0.367 |
|
CINtec+ |
6 (11.32%) |
6 (16.7%) |
5 (55.6%) |
<0.0001 |
|
LSIL |
24 (45.3%) |
22 (61.1%) |
8 (88.9%) |
0.034 |
|
HSIL |
2 (3.8%) |
16 (44.4%) |
5 (55.6%) |
0.000 |
|
ASCUS |
18 (33.9%) |
6 (16.7%) |
3 (33.3%) |
0.185 |
|
NILM |
4 (7.5%) |
0 (0.0%) |
0 (0.0%) |
0.170 |
|
CIN: Cervical Intraepithelial Neoplasia; COC: Combined Oral Contraceptives; STD: Sexually Transmitted Disease; HPV: Human Papillomavirus; HR HPV: High-Risk Human Papillomavirus; HPV 16/18: Human Papillomavirus types 16 and 18; CINtec+: Positive p16/Ki-67 dual staining test (CINtec®); LSIL: Low-grade Squamous Intraepithelial Lesion; HSIL: High-grade Squamous Intraepithelial Lesion; ASCUS: Atypical Squamous Cells of Undetermined Significance; NILM: Negative for Intraepithelial Lesion or Malignancy. |
||||
Table 1: RT-PCR kits used for the diagnosis of COVID-19included in this study.
The distribution of age across the three categories of cervical intraepithelial neoplasia is presented in Figure 1. Among patients with CIN1, the median age was 35 years (interquartile range [IQR], 30–45 years). A similar age distribution was observed in the CIN2 group, with a median age of 34 years and an IQR of 30–45 years. In contrast, patients diagnosed with CIN3 were younger, with a median age of 28 years (IQR, 24–35 years), indicating a shift toward younger age in those with higher-grade lesions. There was no statistically significant difference between groups regarding age distribution (p= 0.12).
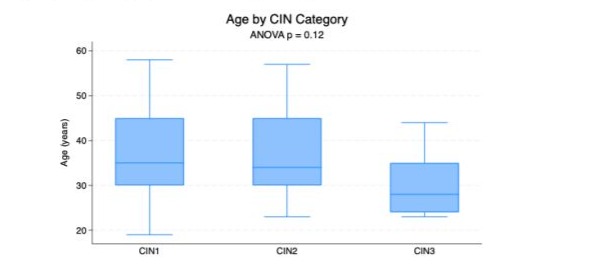
Figure 1: The distribution of age across the three categories of cervical intraepithelial neoplasia (CIN).
The performance metrics of the evaluated algorithms for CIN1 prediction is presented in Table 2 and in Figure 2. In evaluating the performance of machine learning algorithms for the prediction of CIN1, the Random Forest model demonstrated the highest overall performance. It achieved a precision of 0.67, recall of 0.67, and F1-score of 0.67 for the positive class (CIN1), with an overall accuracy of 63% and a ROC AUC of 0.658. The optimized model used no maximum depth restriction and 100 estimators. The K-nearest neighbors (KNN) classifier also showed acceptable performance, with a precision of 0.58, recall of 0.73, and F1-score of 0.65 for CIN1, an accuracy of 56%, and a ROC AUC of 0.539.
Support Vector Machine (SVM) achieved moderate values, with a precision of 0.54, recall of 0.47, and F1-score of 0.50 for the CIN1 group, an accuracy of 48%, and a ROC AUC of 0.444. Logistic regression had a lower performance with a precision of 0.50, recall of 0.40, and F1-score of 0.44 for CIN1, resulting in an overall accuracy of 44% and the lowest ROC AUC of 0.389, despite using a regularization parameter C = 1 and L2 penalty.
Naive Bayes performed modestly with a precision of 0.50, recall of 0.67, and F1-score of 0.57 for CIN1, yielding an accuracy of 44% and ROC AUC of 0.417. While both logistic regression and Naive Bayes offered high recall or precision individually, their overall discrimination was inferior to Random Forest and KNN.
Model | Precision | Recall | F1-score | Accuracy | ROC AUC | Best Parameters |
Logistic Regression | 0.50 | 0.40 | 0.44 | 0.44 | 0.3889 | C = 1, penalty = l2 |
Random Forest | 0.67 | 0.67 | 0.67 | 0.63 | 0.6583 | max_depth = None, n_estimators = 100 |
SVM | 0.54 | 0.47 | 0.50 | 0.48 | 0.4444 | C = 10, kernel = linear |
Naive Bayes | 0.50 | 0.67 | 0.57 | 0.44 | 0.4167 | – |
KNN | 0.58 | 0.73 | 0.65 | 0.56 | 0.5389 | n_neighbors = 5 |
CIN1: Cervical Intraepithelial Neoplasia grade 1; SVM: Support Vector Machine; KNN: k-Nearest Neighbors; ROC AUC: Receiver Operating Characteristic Area Under the Curve; F1-score: Harmonic Mean of Precision and Recall; C: Regularization Parameter; L2: Ridge Regularization Penalty; Max Depth: Maximum Depth of the Trees in Random Forest; n_estimators: Number of Estimators (Trees) in Random Forest | ||||||
Table 2: Performance metrics of machine learning models for CIN1 prediction.
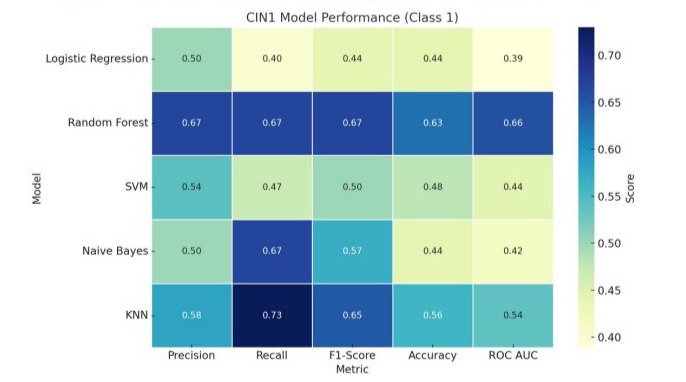
Figure 2: Performance metrics of machine learning models for CIN1 prediction.
The performance metrics of the evaluated algorithms for CIN2 prediction is presented in Table 3 and in Figure 3. Logistic regression with L2 regularization (C = 0.01) exhibited the strongest overall performance, achieving an accuracy of 0.89, a precision of 0.83, a recall of 0.71, and an F1-score of 0.77. Its discriminative ability, as indicated by the ROC AUC, was 0.8393, suggesting a good balance between sensitivity and specificity.
CatBoost, a gradient boosting model, also showed high performance with an accuracy of 0.85, precision and recall both at 0.71, and an F1-score of 0.71. It yielded the highest ROC AUC among all models at 0.8464, indicating excellent predictive power. This model was tuned using a depth of 5 and 100 iterations.
Random Forest, configured with a maximum depth of 10 and 50 estimators, reached an accuracy of 0.78, with precision, recall, and F1-score each at 0.57. Its ROC AUC of 0.8036 indicated solid model discrimination.
The Support Vector Machine with a radial basis function kernel (C = 10) achieved an accuracy of 0.74, precision of 0.50, recall of 0.71, and an F1-score of 0.59, with a ROC AUC of 0.7893. The k-Nearest Neighbors (KNN) model (k = 5) provided similar performance, with an accuracy of 0.78, precision of 0.56, recall of 0.71, and an F1-score of 0.62. Its ROC AUC was 0.7750, indicating moderate predictive power.
XGBoost, another ensemble learning model, achieved an accuracy of 0.74, with precision of 0.50, recall of 0.57, and F1-score of 0.53. Its ROC AUC stood at 0.7964, which was comparable to other ensemble approaches. In contrast, Naive Bayes demonstrated the weakest performance, with an accuracy of 0.37, precision of 0.27, and an F1-score of 0.41, despite a relatively high recall of 0.86. Its ROC AUC was 0.6893, suggesting limited discriminatory capability.
|
Model |
Precision |
Recall |
F1score |
Accuracy |
ROC AUC |
Best Parameters |
|
Logistic Regression |
0.83 |
0.71 |
0.77 |
0.89 |
0.8393 |
C = 0.01, penalty = l2 |
|
Random Forest |
0.57 |
0.57 |
0.57 |
0.78 |
0.8036 |
max_depth = 10, n_estimators = 50 |
|
SVM |
0.50 |
0.71 |
0.59 |
0.74 |
0.7893 |
C = 10, kernel = rbf |
|
Naive Bayes |
0.27 |
0.86 |
0.41 |
0.37 |
0.6893 |
– |
|
KNN |
0.56 |
0.71 |
0.62 |
0.78 |
0.7750 |
n_neighbors = 5 |
|
XGBoost |
0.50 |
0.57 |
0.53 |
0.74 |
0.7964 |
learning_rate = 0.1, max_depth = 5, n_ estimators = 100 |
|
CatBoost |
0.71 |
0.71 |
0.71 |
0.85 |
0.8464 |
depth = 5, iterations = 100 |
|
CIN2: Cervical Intraepithelial Neoplasia grade 2; SVM: Support Vector Machine; KNN: k-Nearest Neighbors; ROC AUC: Receiver Operating Characteristic Area Under the Curve; F1-score: Harmonic Mean of Precision and Recall; C: Regularization Parameter; L2: Ridge Regularization Penalty; Max Depth: Maximum Depth of the Trees in Random Forest; n_estimators: Number of Estimators (Trees) in Random Forest |
||||||
Table 3: Performance metrics of machine learning models for CIN2 prediction.
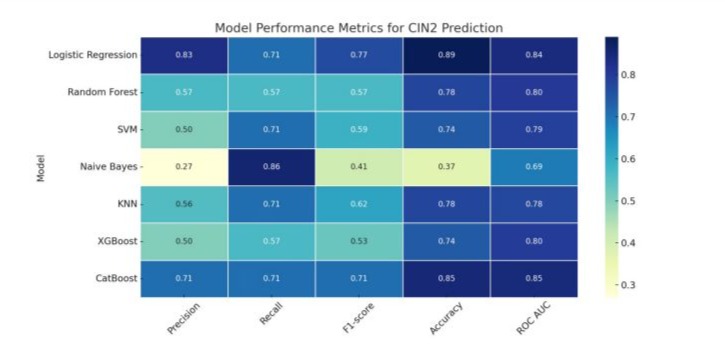
Figure 3: Performance metrics of machine learning models for CIN2 prediction.
The performance metrics of the evaluated algorithms for CIN3 prediction is presented in Table 4 and in Figure 4. Logistic regression demonstrated the highest overall performance among all evaluated models. It achieved a precision of 0.67, perfect recall of 1.00, and an F1-score of 0.80. The model also attained the highest accuracy (96%) and ROC AUC (0.98), with optimal hyperparameters of regularization parameter C = 0.01 and L2 penalty.
Random forest, XGBoost, and CatBoost models all exhibited comparable performance, each reaching an accuracy of 93% and an F1-score of 0.50. These models also showed strong discriminative ability with ROC AUC values of 0.98, 0.93, and 0.96, respectively. The best-performing hyperparameters for random forest included a maximum tree depth of 5 and 50 estimators; XGBoost performed best with a learning rate of 0.05, maximum depth of 3, and 50 estimators; and CatBoost used a depth of 3 and 50 iterations.
The support vector machine model, tuned with C = 0.1 and a linear kernel, showed a precision of 0.33, recall of 0.50, and F1-score of 0.40. While its accuracy was 89%, the ROC AUC was slightly lower at 0.88. K-nearest neighbors (KNN) with 3 neighbors performed less favorably, yielding a precision of 0.25, recall of 0.50, F1-score of 0.33, and accuracy of 85%, with a notably lower ROC AUC of 0.66.
Naive Bayes produced the lowest precision (0.17) but achieved a recall of 1.00, suggesting high sensitivity but poor specificity. Despite a modest F1-score of 0.29, its ROC AUC was high at 0.98, though the overall accuracy remained relatively low at 63%.
Model | Precision | Recall | F1-score | Accuracy | ROC AUC | Best Parameters |
Logistic Regression | 0.67 | 1.0 | 0.8 | 0.96 | 0.98 | = 0.01, penalty = l2 |
Random Forest | 0.5 | 0.5 | 0.5 | 0.93 | 0.98 | max_depth = 5, n_estimators = 50 |
SVM | 0.33 | 0.5 | 0.4 | 0.89 | 0.88 | = 0.1, kernel = linear |
Naive Bayes | 0.17 | 1.0 | 0.29 | 0.63 | 0.98 | - |
KNN | 0.25 | 0.5 | 0.33 | 0.85 | 0.66 | n_neighbors = 3 |
XGBoost | 0.5 | 0.5 | 0.5 | 0.93 | 0.93 | learning_rate = 0.05, max_depth = 3, n_estimators = 50 |
CatBoost | 0.5 | 0.5 | 0.5 | 0.93 | 0.96 | depth = 3, iterations = 50 |
CIN3: Cervical Intraepithelial Neoplasia grade 3; SVM: Support Vector Machine; KNN: k-Nearest Neighbors; ROC AUC: Receiver Operating Characteristic Area Under the Curve; F1-score: Harmonic Mean of Precision and Recall; C: Regularization Parameter; L2: Ridge Regularization Penalty; Max Depth: Maximum Depth of the Trees in Random Forest; n_estimators: Number of Estimators (Trees) in Random Forest | ||||||
Table 4: Performance metrics of machine learning models for CIN3 prediction.
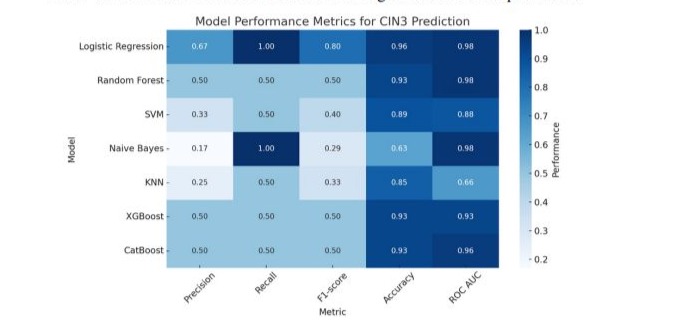
Figure 4: Performance metrics of machine learning models for CIN3 prediction.
Discussion
In this study, we evaluated the performance of multiple supervised machine learning (ML) models in predicting cervical intraepithelial neoplasia (CIN) grades using clinical, cytological, virological, and immunocytochemical data from 98 patients with histologically confirmed CIN1, CIN2, or CIN3.
Consistent with existing literature, our cohort exhibited an increasing prevalence of high-risk HPV types and sexually transmitted infections with higher CIN grades. Notably, HPV 16/18 was found in 88.9% of CIN3 cases, compared with 50% and 22.6% in CIN2 and CIN1, respectively. This finding aligns with regional epidemiologic patterns, as Eastern Europe continues to report some of the highest HPV prevalence and dysplasia rates globally. A recent 10-year epidemiological study from Serbia reported a strikingly high overall HPV positivity rate of 43.3% among women, highlighting the widespread nature of infection in the region. Among the detected HPV strains, high-risk genotypes, including HPV types 16, 31, 52, 56, 39, and 51, accounted for 62.3% of infections. Notably, HR HPV was detected in 76.5% of women diagnosed with HSIL [16]. At a broader regional level, Central and Eastern Europe (CEE) continues to experience a substantial burden of HPV-related diseases. In 2019 alone, across nine CEE countries, including Bulgaria, Croatia, Czechia, Hungary, Poland, Romania, Serbia, Slovakia, and Slovenia, there were an estimated 6,832 deaths attributable to HPV-related cancers, alongside 107,846 years of life lost [17]. These statistics reflect both a high prevalence of oncogenic HPV infection and substantial progression rates to dysplasia and malignancy.
CINtec® dual-staining demonstrated strong association with lesion severity in our cohort. While 55.6% of CIN3 cases were CINtec-positive, the positivity declined to 16.7% in CIN2 and just 11.32% in CIN1. Studies consistently demonstrate that CINtec® dual-stain positivity correlates strongly with lesion severity. In particular, the proportion of dual-stain–positive samples increases progressively across CIN grades: approximately 50% in lesions classified as ≤CIN1, 76.6%–100% in CIN2, and 87%–100% in CIN3 [18-20]. In cases of invasive cervical cancer, dual-stain positivity remains high, reaching 83.3% [18]. These findings suggest a robust quantitative association between p16/Ki-67 coexpression and histological severity, reinforcing the role of dualstaining as a reliable biomarker for high-grade disease [18, 19, 21].
The density of dual-positive cells is significantly elevated in CIN2 and CIN3 lesions compared to low-grade or benign findings, further supporting its biological relevance as an indicator of transforming HPV infections [19, 21]. Moreover, when compared with HPV genotyping or cytological assessment alone, dual-staining offers superior diagnostic specificity and overall accuracy for detecting CIN2+ lesions, with reported performance metrics reaching 83.5% for accuracy and 84.8% for specificity [20, 22, 23].
From a clinical standpoint, these findings highlight the utility of dual-staining in triaging equivocal cytology results and identifying women at increased risk for high-grade lesions. For instance, in patients with CIN2, dual-stain positivity ranges from 76.6% to 100%, while in CIN3 cases it often approaches or exceeds 87%, offering a stratified risk framework to guide further intervention [18, 19, 21]. In contrast, lesions classified as ≤CIN1 typically demonstrate lower rates of positivity (~50%), suggesting that a negative result may help support conservative management strategies [19].
The ML models demonstrated variable predictive power depending on the CIN grade and underlying algorithm. For CIN1 prediction, Random Forest and KNN models outperformed others, with F1-scores of 0.67 and 0.65, respectively. These models may have benefited from their capacity to handle mixed-type data and capture non-linear patterns in clinical and behavioural variables, such as number of sexual partners and smoking history.
In contrast, CIN2 prediction was best achieved using Logistic Regression and CatBoost, with F1-scores of 0.77 and 0.71, respectively. Logistic regression achieved the highest accuracy (89%) and a robust ROC AUC (0.8393), suggesting that linear decision boundaries combined with regularization may be sufficient for moderate-grade lesions, particularly when guided by strong predictors like HPV 16/18 status and HSIL cytology.
CIN3 prediction was the most accurate across all models. Logistic Regression again led performance with an F1-score of 0.80 and an high AUC value of 0.98. Other ensemble methods such as Random Forest, XGBoost, and CatBoost also performed well (accuracy ≥ 93%, ROC AUC ≥ 0.93). Despite the small number of CIN3 cases (n=9), these results highlight the ability of ML models to detect high-risk lesions with strong discriminative power, a critical need in cervical cancer prevention programs. Notably, while Naive Bayes models frequently achieved high recall, they consistently underperformed in precision and overall accuracy, particularly in CIN3 (precision = 0.17). This reflects the model’s sensitivity to assumptions of feature independence, which are often violated in real-world medical datasets.
Recent advancements in machine learning have shown the efficacy of different algorithms in predicting cervical intraepithelial neoplasia. Traditional models like logistic regression and KNN are widely utilized for their interpretability and simplicity; however, they are often surpassed by ensemble and advanced models such as Random Forest, XGBoost, and CatBoost, particularly when applied to high-quality, pre-processed clinical data [24-27].
SVMs demonstrated notable efficacy in cervical pathology tasks, with AUC values reported between 0.82 and 0.93 in diagnostic applications, including degenerative cervical myelopathy and cervical cancer screening [24, 25]. While SVMs do not consistently outperform ensemble methods, they are a reliable option for binary classification tasks involving balanced datasets.
Ensemble techniques, especially stacked ensembles, demonstrate superior predictive performance. A study on cervical cancer detection demonstrated that a stacked ensemble model attained an accuracy of 0.994, markedly surpassing the performance of individual classifiers [5]. Random Forest models, although not consistently evaluated for CIN1–3 stratification, have demonstrated robust performance in broader cervical neoplasia prediction tasks and are often preferred for their capacity to manage imbalanced and non-linear data [26].
KNN, while employed in various studies mainly for imputation or as a secondary classifier, generally demonstrates lower predictive accuracy and reduced robustness in comparison to ensemble or kernel-based models [27]. Similarly, although XGBoost and CatBoost are commonly utilized in medical machine learning tasks, their specific application to CIN prediction is not well-documented in the existing literature. Their comparative performance in CIN grading tasks is largely speculative and requires further investigation.
In summary, although ensemble and advanced machine learning models show significant potential for accurate prediction of CIN grades, existing literature does not provide direct, comparative analyses of these algorithms within this particular diagnostic framework [26, 27]. Most existing studies primarily address cervical cancer or general neoplasia, rather than specifically examining stratified CIN grading. Further research is required to establish benchmarks for these models in predicting CIN1, CIN2, and CIN3, utilizing standardized datasets and evaluation metrics.
This study has several limitations. First, the small sample size, especially the small number of CIN3 cases, may have influenced model stability and generalizability. Although SMOTE was used to mitigate class imbalance, synthetic oversampling cannot fully replicate the complexity of naturally occurring high-grade lesions. Second, our dataset was derived from a single geographical region, which may limit external validity. Differences in HPV genotypes, screening coverage, and healthcare infrastructure may influence model applicability across populations. Third, the study employed classical ML approaches rather than end-to-end deep learning pipelines, which could potentially further improve classification if larger annotated image datasets were available.
Future research should explore larger, multicenter datasets incorporating multimodal inputs, including colposcopic imagery, digital pathology, and molecular biomarkers. Additionally, prospective validation of the best-performing models is necessary to evaluate clinical utility. Integrating ML predictions with clinical decision support systems could offer tailored risk stratification, thereby improving adherence to guideline-based management.
Conclusion
Our findings highlight the significance of high-risk HPV genotyping, particularly types 16 and 18, along with CINtec® dualstaining, as essential factors in predicting CIN. CINtec positivity demonstrated a significant correlation with lesion severity, thereby supporting its function as a surrogate biomarker for oncogenic transformation.
While the study presents encouraging results, limitations including a small sample size and geographic specificity necessitate caution in generalizing these findings. Despite class imbalance, the consistently high accuracy in CIN3 detection highlights the potential of machine learning models to enhance early cervical cancer risk assessment and management.
Subsequent research should focus on validating these findings in larger, multicenter cohorts, preferably integrating multimodal data, including colposcopic imaging and molecular biomarkers. Furthermore, the clinical integration of machine learningdriven tools should prioritize the improvement of decisionmaking workflows, the reduction of diagnostic variability, and the mitigation of both over-treatment and under-treatment of precancerous cervical lesions.
Author Contribution Statement
Designed research: I.V.M, D.S, R.S, I.-A.V, A.-M.C. Performed research: I.V.M, A.-M.A, G.A, P.V, V.H, A.H. Analyzed data: I.V.M, I.-A.V, A.-M.C. Data acquisition: A.-M.A, G.A, P.V.
Writing—original draft: I.V.M, A.-M.C. Review & editing: All authors. All authors have read and agreed to the published version of the manuscript.
Ethics Approval Statement
The protocol for this research project has been approved by the Institutional Ethics Committee of Clinical Hospital of Obstetrics and Gynecology Buna vestire” Galati (No. 115/05.01.2021) and it conforms to the provisions of the Declaration of Helsinki.
Funding
This work received no external funding.
Data Availability Statement
Data generated during this study are available from the corresponding author on reasonable request.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- Javanbakht Z, Kamravamanesh M, Rasulehvandi R, Heidary A, Haydari M (2023) Global Prevalence of Cervical Dysplasia: A Systematic Review and Meta‐Analysis. Indian Journal of Gynecologic Oncology 21.
- Forman D, De Martel C, Lacey C, Soerjomataram I, Lortet-Tieulent J, et al. (2012) Global burden of human papillomavirus and related diseases. Vaccine 30 Suppl 5.
- Poljak M, Seme K, Maver P, Kocjan B, Cuschieri K, et al. (2013 Human papillomavirus prevalence and type-distribution, cervical cancer screening practices and current status of vaccination implementation in Central and Eastern Europe. Vaccine. 31 Suppl 7.
- Ismail S, Colclough A, Dinnen J, Eakins D, Evans D, et al. (1989) Observer variation in histopathological diagnosis and grading of cervical intraepithelial neoplasia. British Medical Journal 298: 707-10.
- Masters N (1989) Grading of cervical intraepithelial neoplasia. British Medical Journal. 298:1031.
- Jenkins D, Jarmulowicz M (1989) Grading of cervical intraepithelial neoplasia. British Medical Journal 298: 1030-1.
- Scripcariu I-S, Gisca T, Botezatu A, Socolov D, Vasilache I-A, et al. (2025) Evaluating the Predictive Performance of miR-124-2 and FAM19A4 for Cervical Lesions in a Single Center from Romania: A Prospective Study. Journal of Clinical Medicine 14: 3452.
- Gisca T, Munteanu IV, Vasilache IA, Melinte-Popescu AS, Volovat S, Scripcariu IS, et al. (2024) A Prospective Study on the Progression, Recurrence, and Regression of Cervical Lesions: Assessing Various Screening Approaches. J Clin Med 13(5).
- SC-SENet: A Classification Diagnosis Model for Cervical Intraepithelial Neoplasia Based on Colposcopic Images. Modeling and Simulation 2025.
- Brosnan B, Skarga-Bandurova I, Biloborodova T, Skarha-Bandurov I (2023) An Integrated Approach to Automated Diagnosis of Cervical Intraepithelial Neoplasia in Digital Histology Images. Studies in health technology and informatics 302: 615-6.
- Brosnan B, Skarga-Bandurova I, Biloborodova T, Skarha-Bandurov I (2023) Cervical Intraepithelial Neoplasia Grading from Prepared Digital Histology Images. Studies in health technology and informatics 305: 402-5.
- Mukku L, Thomas J (2024) CMT-CNN: colposcopic multimodal temporal hybrid deep learning model to detect cervical intraepithelial neoplasia. International Journal of Advances in Intelligent Informatics.
- An H, Ding L, Mengyuan, Huang A-H, Gan Y, et al. (2023) Deep Learning-Based Recognition of Cervical Squamous Interepithelial Lesions. Diagnostics 13.
- Yang Y, Zhou H, Wang Y, Dai Y, Pi R, et al. (2025) [Construction and preliminary validation of machine learning predictive models for cervical cancer screening based on human DNA methylation]. Zhonghua zhong liu za zhi [Chinese journal of oncology] 47 :193-200.
- Liu L, Liu J, Su Q, Chu Y, Xia H, et al. (2024) Performance of artificial intelligence for diagnosing cervical intraepithelial neoplasia and cervical cancer: a systematic review and meta-analysis. eClinicalMedicine 80.
- Nikolić N, Bašica B, Štrbac M, Terzic L, Patić A, et al. (2024) Prevalence of Carcinogenic Genotypes of HPV-Infected Women in a Ten-Year Period (2014–2023) in Vojvodina, Serbia. Medicina 60.
- Sabale U, Reuschenbach M, Takyar J, Dhawan A, Hall A, et al. (2025) Epidemiological, economic and humanistic burden of cervical intraepithelial neoplasia in Europe: A systematic literature review. European Journal of Obstetrics & Gynecology and Reproductive Biology: X 25:100360.
- Van Der Merwe H, Dreyer G, Visser C, Botha M, Whittaker J, Snyman L, et al. (2024) 66 Test performance of dual stain cytology in a cervical screening population with high prevalence of HIV and precursor lesions. Poster and E-Posters.
- Hashimoto K, Kumagai T, Nomura K, Miyagawa Y, Tago S, et al. (2023) Validation of an on-chip p16ink4a/Ki-67 dual immunostaining cervical cytology system using microfluidic device technology. Scientific Reports 13.
- Kakaliashvili-Dzagnidze S, Khardzeishvili O, Tabagari S (2023) Diagnostic Accuracy of p16INK4a/Ki-67 Dual Immunostaining for Detection of High-Grade Cervical Intraepithelial Neoplasia in Women Involved in Cervical Cancer Screening in Georgia. Analytical Cellular Pathology (Amsterdam) 2023.
- Kolotusha V, Khoperiya V (2005) Approach to P16 and Ki-67 in the Cervical Intraepithelial Neoplasia staging. The Ukrainian Scientific Medical Youth Journal.
- Gandhi G, Chadha S, Gupta R, Hedau S, Agarwal K (2023) #916 Human papilloma virus 16/18 genotyping with dual staining for the detection of high grade cervical intraepithelial neoplasia in women with low grade cytology. Oral Sessions.
- Bedón AA, Delgado-López D, Ochoa-Avilés C, Rivas-Párraga R, Crespo BV, Verhoeven V, et al. (2024) p16/Ki-67 dual staining as a predictive value for cervical cancer compared to other conventional triage tools: a descriptive literature review. European Journal of Gynaecological Oncology.
- Song J, Li J, Zhao R, Chu X (2023) Developing Predictive Models for Surgical Outcomes in Patients with Degenerative Cervical Myelopathy: A Comparison of Statistical and Machine Learning Approaches. The spine journal : official journal of the North American Spine Society.
- Chu X, Song J, Wang J, Kang H (2025) A stacking ensemble machine learning model for predicting postoperative axial pain intensity in patients with degenerative cervical myelopathy. Scientific Reports 15.
- Nakajo M, Jinguji M, Tani A, Yano E, Hoo CK, et al. (2021) Machine learning based evaluation of clinical and pretreatment 18F-FDG-PET/ CT radiomic features to predict prognosis of cervical cancer patients. Abdominal Radiology 47:838-47.
- Aljrees T (2024) Improving prediction of cervical cancer using KNN imputer and multi-model ensemble learning. PLOS ONE 19.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.