Tumor Cells Identified by Single-Cell RNA Sequencing in A Patient with Nasopharyngeal Mass: A Case Report
by Zhang S, Wen Y, Zheng J*
Department of Radiotherapy, General Hospital of Southern Theatre Command, Guangzhou, China.
*Corresponding author: Zheng J, Department of Radiotherapy, General Hospital of Southern Theatre Command, Guangzhou, China.
Received Date: 28 July, 2024
Accepted Date: 25 September, 2024
Published Date: 27 September, 2024
Citation: Zhang S, Wen Y, Zheng J (2024) Tumor cells identified by single-cell RNA sequencing in a patient with nasopharyngeal mass: a case report. J Oncol Res Ther 9: 10250. https://doi.org/10.29011/2574-710X.10250.
Abstract
Nasopharyngeal cancer is endemic in South China. Early diagnosis and treatment are important for the prognosis of patients. For patients who could not be pathologically diagnosed for the lack of tumor cells in biopsy specimen, treatment might be delayed. Here we report a male patient from endemic region of nasopharyngeal carcinoma with a mass in nasopharynx on magnetic resonance imaging and elevated blood Epstein-Barr virus DNA copies. Nasopharyngeal biopsies were repeated for three times while the pathological examination only showed inflammation of the squamous epithelium with few variant cells. Single-cell RNA sequencing was performed with biopsy tissues and only nine tumor cells were recognized. The patient was pathologically confirmed as undifferentiated non-keratinizing carcinoma several months later. We suspect that single-cell RNA sequencing may contribute to the early diagnosis in cases with very few tumor cells.
Keywords: Nasopharyngeal Carcinoma; scRNA-seq; Diagnosis; Case Report.
Background
Nasopharyngeal carcinoma (NPC) is pandemic in South China, and the incidence in Guangdong province exceeds 10/100,000 [1]. NPC originates from epithelial cells of the nasopharyngeal mucosal lining and is often observed at the pharyngeal recess [2]. In endemic regions, non-keratinizing carcinoma comprises over 95% cases and is associated with Epstein-Barr virus (EBV) [3]. With the development of radiotherapy and medicine approaches, the prognosis of patients with NPC is improving, while the 5-year overall survival rate decreases obviously from 90% for patients at stage I to 58% for those at stage IV [4]. Therefore, early diagnosis and treatment are important to improve survival for patients with NPC. Biopsy of nasopharynx or fine-needle aspiration of the neck nodes are the most common methods to obtain tissues for pathological diagnosis, which is the golden standard for tumor diagnosis. The lack of valid tumor tissues may hurdle pathological diagnosis, delay the timing of treatment, leading to deteriorated survival of patients.
In recent years, with the development of sequencing technologies, single-cell RNA sequencing (scRNA-seq) has progressed rapidly and can offer comprehensive analysis of thousands of individual cells simultaneously, including transcriptional, genomic, epigenomic characteristics. Therefore, it can be used to identify rare subpopulations and different states of tumor cells and immune cells in the tumor microenvironment (TME), improve the understanding of cancer diagnosis, biomarker, treatment and prognosis [6]. Due to the high cost, scRNA-seq has not been widely used in clinic. Here we present a case with masses in the nasopharynx considered as NPC while tumor cells could not be found by pathological examinations. In scRNA-seq analysis, several tumor cells were identified as a subpopulation of epithelial cells. We suspected that scRNA-seq may aid the diagnosis of tumor for such cases.
Case Presentation
A 53-year old male was hospitalized in our department in August 2021. He had experienced nasal congestion for six months, and the symptom was progressively exacerbated. Magnetic resonance imaging (MRI) showed a mass in the nasopharynx involving bilateral staphylinus externus and petrostaphylinus (Figure 1), accompanied with slightly enlarged bilateral level IIA lymph nodes in the neck. 18F-fluorodeoxy-glucose (18FDP)-PET/CT showed mildly increased glucose metabolism of both the primary mass and cervical nodes. Laboratory tests showed EBV DNA copies of 769 (higher than the normal value of 400 in our hospital) in peripheral blood. Nasopharyngoscopy showed an eminence lesion in the nasopharynx, while tumor cells were not found in the biopsy specimen. Repeated nasopharyngeal biopsies were conducted for two more times while histopathological examination only showed acute and chronic inflammation of the squamous epithelium with few variant cells, hyperplasia of lymphatic tissue and minute vessels in submucosa. The diagnosis could not be pathological confirmed while the patient was highly suspected of NPC with the symptom of nasal congestion, the radiological manifestations and elevated EBV DNA copies. Furthermore, he was from Sihui city where the incidence of NPC reached 20.91/100,000 [1].
At the third time of nasopharyngeal biopsy, the patient was informed with a study analyzing the tumor microenvironment of NPC by scRNA-seq in our department. He signed written consent to donate biopsy tissues for scientific research. With the specimen, scRNA-seq analysis was performed including 4634 cells. The most abundant components were B cells (1487), T cells (1722), epithelial cells (467), plasma cells (358), macrophages (198) and dendritic cells (125). In the epithelial cells, only 9 (1.9%) were identified as tumor cells. The number and proportion of tumor cells in the specimen of this patient are listed in Table 1, which are compared with the results from other two patients (P1 and P2) with pathologically confirmed NPC.
The epithelial cells were partitioned into eight sub-clusters based on t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm (Figure 2). The tumor cell cluster was determined by high expression of markers such as STMN1, CCL20, DAPL1 and GPNMB (Figure 3). The tumor cells separated three clusters by the gene expression states, suggesting intra-tumor heterogeneity of nasopharyngeal carcinoma. While in the current case (P3), only one cluster of tumor cells was identified (Figure 4), which may be due to the small amount of tumor cells. Heatmap showed distinct gene expression profile in each cluster of tumor cells (Figure 5). The tumor cells from the current case express HMGB2, TOP2A, PCLAF, CENFB, UBE2C at a high level.
The patient was discharged for the negative pathological results. He underwent regular inspections and was diagnosed as undifferentiated non-keratinizing carcinoma by repeated nasopharyngeal biopsy several months later. He received concurrent chemoradiotherapy and achieved complete response. Follow-up by telephone was continued, and the patient was alive without disease recurrence until May 2024.
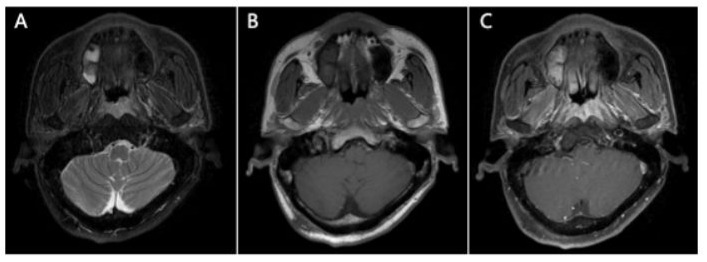
Figure 1: MRI scan showed an apparent contrast-enhancing soft tissue lesion in the nasopharynx involving bilateral staphylinus externus and petrostaphylinus (A) T2-weighted image of the lesion (B) T1-weighted image (C) contrast-enhanced T1-weighted image.
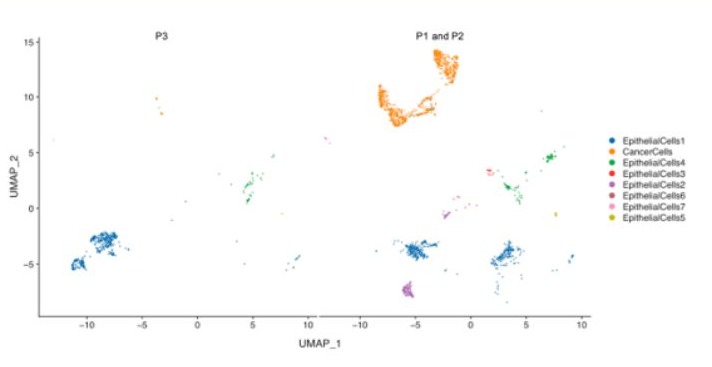
Figure 2: t-SNE plot of epithelial cells from the biopsy tissues of the current case (P3) and other two patients (P1 and P2) diagnosed as undifferentiated non-keratinizing carcinoma. The subtypes of epithelial cells are roughly consistent among patients.
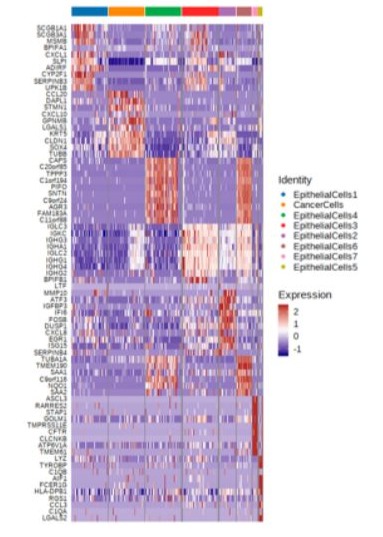
Figure 3: Heat map of differentially expressed genes (rows) in the eight clusters of epithelial cells from the three patients. The top ten genes overexpressed in each cluster are indicated.
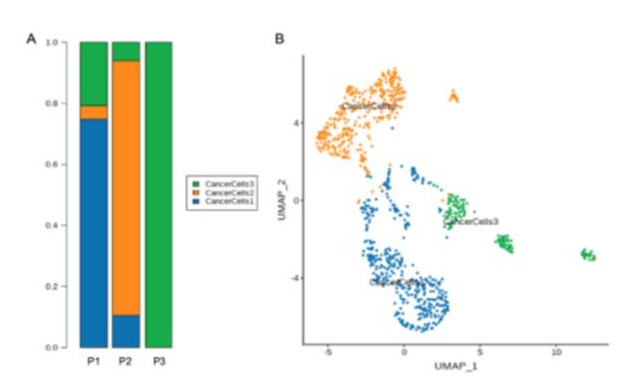
Figure 4: (A)The proportion and composition of tumor cell clusters among three patients. (B) Tumor cells can be classified into three clusters and are annotated as cancer cells 1, cancer cells 2 and cancer cells 3. All of the three clusters are identified from P1 and P2, while only cluster 3 is identified from the current case (P3).
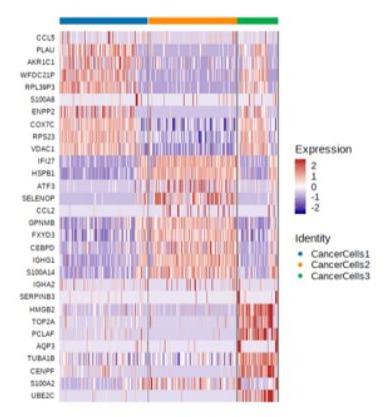
Figure 5: Heat map of differentially expressed genes (rows) in the three clusters of tumor cells. The three clusters of tumor cells have different individual expression patterns. The tumor cells identified from this case belong to the cluster of CancerCells 3.
|
All cells |
Epithelial cells |
Tumor cells |
|
|
Patient 1 |
5331 |
1058 |
474 |
|
Patient 2 |
6833 |
560 |
389 |
|
Patients 3 (the current case) |
5159 |
467 |
9 |
Table 1: The number of epithelial cells and tumor cells identified in the biopsy tissues.
Discussion
There were few reports on diagnosis of tumor with scRNA-seq. In this case, repeated biopsies were conducted while tumor cells were not found with conventional pathological examination. Tissue biopsy is invasive, and the accuracy is affected by many factors such as the skills of the operator, the distribution of tumor cells. Repeated biopsies may increase the risk of bleeding and infection, therefore, techniques that can help to find tumor cells are needed under such circumstances. The failure of pathological diagnosis in this case may be caused by the scarcity of tumor cells, as only nine tumor cells were identified by scRNA-seq. Tumor cells proliferated gradually and accumulated to an enough amount to be confirmed by pathological examination several months later. The subsequent pathology verified the accurate finding of tumor cells by scRNA-seq, indicating the value of this technique in early diagnosis of tumor. Currently, pathology is still the golden standard for tumor diagnosis, we could not start treatment only according to tumor cells identified by scRNA-seq. Fortunately, the disease did not progress significantly and was cured preliminarily in this case, while for patients with more aggressive tumors, the delay of pathological diagnosis may deprive the opportunity for effective treatment.
In endemic regions, NPC is closely associated with EBV infection. Genetic alterations in premalignant nasopharyngeal epithelium, inflammatory stimulation in the nasopharyngeal mucosa allow the infection of EBV to be latent. During latent infection, the expression product of viral genes drives premalignant epithelial cells into tumor cells [6]. In this case, the very few tumors cells may indicate that the lesion of this patient was in a stage of malignant transformation. Individuals with latent EBV infection may mount anti-viral immune response to restrict the number of infected cells, which gives the TME an inflammatory appearance. At the same time, there are local immunosuppressive factors which facilitate tumor development against anti-viral immune response. Previous studies found that the TME of EBV+ nasopharyngeal were heavily infiltrated with immune cells including T cells , B cells, natural killer cells, dendritic cells, macrophages and so on, and the composition of TME varied among individual cases [7], which were consistent with the findings in our case. The overexpressed genes of tumors cells found in this case were also reported in previous studies on NPC. HMGB2 may relate to the immunosuppressive TME [8]. PCLAF was reported to be a key gene contributing to the genesis and progression in NPC [9]. Overexpression of TOP2A was found to contribute to tumor aggressiveness in a subset of NPC [10]. High UBE2C expression level related to clinical progression of NPC and may be a potential target for treatment [11]. One advantage of scRNA-seq is the comprehensive annotations of gene expressions. Further studies on the gene expression status and interplay of various cells a in the TME may advance the knowledge on the carcinogenic process and provide potential biomarkers for treatment.
There are limitations for our work. Firstly, this is a case report, and the feasibility of scRNA-seq in tumor diagnosis should be verified in more cases. Secondly, scRNA-seq revealed the transcriptional profile of tumor cells, while the function of abnormally expressed genes need further demonstrations. Thirdly, the characteristics of immune cells, the interplay between tumor cells, immune cells and EBV need further illustration.
Conclusion
In conclusion, given the failure of conventional pathological examination, scRNA-seq might provide a precise approach to find tumor cells and assist in diagnosis.
Acknowledgment: We acknowledge the patient for the agreement of publication of the clinical, radiological and sequencing data. This work was supported by the Science and Technology Planning Projects of Guangzhou (Grant No. 202102021259).
Declaration of conflict of interest: There were no conflicts of interest.
References
- Wei KR, Zheng RS, Zhang SW, Liang ZH, Li ZM et al. (2017) Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chin J Cancer 36:90.
- Su ZY, Siak PY, Lwin YY, Cheah SC (2024) Epidemiology of nasopharyngeal carcinoma: current insights and future outlook. Cancer Metastasis Rev 43:919-939.
- Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, et al. (2019) Nasopharyngeal carcinoma. Lancet 394:64-80.
- Lee AW, Sze WM, Au JS, Leung T W, Chua D T T, et al. (2005) Treatment results for nasopharyngeal carcinoma in the modern era: the Hong Kong experience. Int J Radiat Oncol Biol Phys 61:1107-1116.
- Lei Y, Tang R, Xu J (2021) Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol 14:91.
- Tsang CM, Tsao SW (2015) The role of Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Virol Sin 30:107-121.
- Tan GW, Visser L, Tan LP, Van den Berg A, Diepstra A (2018) The Microenvironment in Epstein-Barr Virus-Associated Malignancies. Pathogens 7:40.
- Neubert EN, DeRogatis JM, Lewis SA (2023) HMGB2 regulates the differentiation and stemness of exhausted CD8+ T cells during chronic viral infection and cancer. Nat Commun 14:5631.
- Ma F, Zhi C, Wang M (2020) Dysregulated NF-κB signal promotes the hub gene PCLAF expression to facilitate nasopharyngeal carcinoma proliferation and metastasis. Biomed Pharmacother 125:109905.
- Lan J, Huang HY, Lee SW (2014) TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol 35:179-187.
- Shen Z, Jiang X, Zeng C (2013) High expression of ubiquitin-conjugating enzyme 2C (UBE2C) correlates with nasopharyngeal carcinoma progression. BMC Cancer 13:192.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.