Study to Identify the Signalling Cascade behind Expression Level of PTEN and RB1 gene in Breast Cancer
Haris Abdul Rehman1,Mah Noor Hassan2, Muhammad Umar3, Aqsa Qurban4*, Hafiz Khawar5,Iqra Jamil6,Najeeb Ullah Khan7,Syeda Anamta Hashmi8,Fidaa Aslam9
1Department of Microbiology, University of Central Punjab, Lahore-54000, Pakistan
2Department of Biochemistry, University of Central Punjab, Lahore-54000, Pakistan
3Department of Biochemistry, Government College Women University Faisalabad-38000, Pakistan
4Department of Life Sciences, University of Management and Technology, Lahore-54000, Pakistan
5Department of Biotechnology, Government College University Lahore-54000, Pakistan
6Department of Medical Laboratory sciences, Government College University Lahore-54000, Pakistan
7Institue of Biotechnology and Genetic Engineering, The University of Agriculture Peshawar-25000, Pakistan
8Department of Microbiology, University of Lahore-54000, Pakistan
9Centre of Excellence in Molecular Biology, Punjab University Lahore-54000, Pakistan
*Corresponding author: Aqsa Qurban, Department of Life Sciences, University of Management and Technology, Lahore-54000, Pakistan
Received Date: 15 August, 2022
Accepted Date: 07September, 2022
Published Date: 13September, 2022
Citation: Rehman HA, Hassan MN, Umar M, Qurban A, Khawar H, et al. (2022) Study to Identify the Signalling Cascade behind Expression Level of PTEN and RB1 gene in Breast Cancer.Rep GlobHealth Res 5: 142. DOI: https://doi.org/10.29011/2690-9480.100142
Abstract
Breast cancer is the second most prevalent cancer among Pakistani women and the disease burden of breast cancer is continuously uprising. There are several genes involved in breast cancer incidence. The current study was designed to analyze the gene expression level of hereditary onco-suppressive PTEN and RB1, proto-oncogenes i.e. Src and KRAS, and microRNA 140/145/238in breast cancer patients performed through qRT-PCR.Results showed significant up-regulation of proto-oncoSrc and KRAS genes (p<0.05). On the other hand, a significant down-regulation of hereditary onco-suppressive PTEN and RB1 genes while as, the micro RNA signalling cascade involvement through higher expression levels of MI-140, Mi-145, and Mi-238 was also observed (p<0.05). Biopsy samples were preserved in 10% formalin for histopathology as well as in Trizol for mRNA extraction. Histopathlogical examination showed multilayering, hyperplasia, and a complete distortion of ductal and glandular epithelium of breast gland in breast cancer patients.
Keywords: Breast cancer; Histopathology; Proto-oncogenes; Down regulation; mRNA signalling; Expression level
Introduction
Pakistan is ranked highest for mortality and incidence of breast cancer[1-3]. According to 2020 statistics, 2.3 million women were reported with an incidence of breast cancer with 685,000 deaths [4].In 2020, the detailed breast cancer cases in Pakistan were 25,928 which represented 14.5% of a wide range of diseases [4].
Genes associated with breast cancer incidence include BRCA1, BRCA2, PALB2 (Partner And Localizer of BRCA2), CHEK2 (Checkpoint Kinase 2), CDH1 (CaDHerin 1), PTEN (Phosphatase and TENsin homolog), STK11 (Serine/Threonine Kinase 11), Tp53 (Tumor Protein p53), RB1, src, KRAS, ATM, BARD1, BRIP1, CASP8, CTLA4, CYP19A1, FGFR2, H19, LSP1, MAP3K1, MRE11A, NBN, RAD51, and TERT[5,6]. Oncogenesis mainly contributed to mutations in two types of genes: tumor suppressor genes and proto-oncogenes[7]. PTEN and RB1 are examples of tumor suppressor genes while Src and KRAS belong to a class of proto-oncogenes.
PTEN is a tensin and phosphatase homolog that on chromosomal deletion on chromosome number 10.PTEN plays an important part not only in apoptosis induction and arrest of the cell cycle but also in many other physiological functions including migration, differentiation, and cellular adhesion.PTEN is an important tumor-suppressing gene in breast cancer and normally suppresses cellular proliferation by down-regulating the PI3K/ AKT signalling pathway.mTOR/PI3K/AKT is the most commonly deregulated pathway in a different type of carcinogenesis including breast cancer[8]. PTEN is one of the most commonly mutated tumor suppressor genes in human malignancies. Many different types of mutations that occur at the genetic level have been found to occur at phosphatase and tensin homolog including insertions, deletions, frameshift mutations, splice site variants, nonsense, and missense mutations that are linked with the cancers associated with complete inhibition or reduction in phosphatase activity of the phosphatase and tens in homolog deleted on chromosome ten.
RB1 is retinoblastoma which is normally controlling many important physiological functions of the cell including the formation of retinoblastoma protein, cell survival, cell cycle progression, control of apoptosis, or programmed cell death. The transcription of early growth factor-2 receptor is inhibited by the canonical type retinoblastoma protein. The retinoblastoma protein normally functions in signaling pathways that are controlled by its most important regulators working in the upstream direction including CDK4, p16, and cyclin D1 which controls the activation of retinoblastoma protein via its phosphorylation and also controls the ability of retinoblastoma protein to inhibit the normal
functioning of early growth factor 2 receptors [8]. Many different types of mutations are reported at various steps of retinoblastoma signalling pathways in various types of cancers reported to date. RB1 is normally a tumor suppressor gene, loss of expression of RB1 results in the development of basal-like breast cancer[9].
Src, the gene product of avian Rous sarcoma virus is reported for progression, development, and maintenance of various forms of cancer including breast cancer is a proto-oncogene belonging to Src family kinases (SFKs)[10]. Src is an oncogenic steroid receptor coactivator. It is reported that alterations in type enzymes controlling two types of functions including transcriptional programming, and cellular metabolism are one of the most important hallmarks of cancer resulting in uncontrolled proliferation and metastasis[11]. According to one study, the metabolic enzyme(PFKB4) regulates the transcriptional factor responsible by activating the Src, and any type of mutation in that enzyme results in a mutational change in Src, and as a result, there will be initiation and progression of carcinogenesis including breast cancer. Studies report the decrease of invasion and proliferation of cancer cells when Src was disrupted genetically[10].
KRAS (Kirsten rat sarcoma virus) proto-oncogene located on 12p12.1 belongs to the RAS superfamily of small GTPases and is involved in the RAS/MAPK pathway to relay signals for cellular proliferation and growth[12]. The key feature of the RAS family is the presence of catalytic G- domain. The most frequently studied proteins in the RAS subfamily include K-RAS (KRAS4A and KRAS4B, N-RAS, and H-RAS[13]. KRAS is usually associated with the progression of the cell cycle while in cases of increased levels can also induce apoptosis and growth arrest[14]. Wild-type KRAS is usually associated with tumor suppression while the mutated version of genes induces oncogenic properties. KRAS is one of the most commonly mutated proto-oncogenes in human breast cancers most frequently in triple-negative breast cancer[15]. Most of the studies indicated that functions of KRAS in normal cells are controlled mainly by the miRNAs. In the case of triple negative breast cancer one of the mutations in miRNA-873 results in loss of the normal functional ability of KRAS and as a result, there will be initiation and progression of carcinogenesis in various parts of the body including breast tissue causing breast carcinogenesis. Oncogenic KRAS mutations are seen in a round of about 15% of all tumors and deregulation of the MEK/RAF/ ERK pathway by extracellular signal-regulated kinase (KRAS) hyperactivation is found in roughly 30% of all tumors.
MicroRNAs are a group of small endogenous non-coding RNAs that impart a significant role in controlling the gene expression level in normal body cells. Many pieces of evidence indicated that disorganized expression of microRNAs occurs in various types of cancers through different types of mechanisms including microRNA genes amplification or deletion, dysregulated
control of microRNA at the transcriptional level, unorganized changes at the epigenetic level, and defects at different levels of biogenesis networking of microRNAs.miRNAs play a crucial part in the development and proliferation of pathogenesis of solid tumors and assist in their role as tumor suppressors and proto
oncogenes[16]. In the diffused form of B cell large lymphoma elevated expression of unorganized and abnormal microRNAs was identified in serum samples. Some non-coding large microRNAs also play a crucial part in the progression and development of breast cancer by altering its regulatory functions through various mechanisms including interaction with different types of proteins including modifiers of the epigenetic system, and transcriptional co-activators [17].
In this study, we analyzed the expression of miRNA-140, mi-RNA145, miRNA-238, PTEN, RBA1, KRAS, and Src in breast cancer patients and control along with the Histopathological Examination of adenocarcinoma, ductal carcinoma, fibrous carcinoma, and lobular carcinoma in breast tissues.
Methodology
Sample Collection
The clinical study was conducted on breast cancer patients of Allied Hospital Faisalabad with the permission of the ethical review committee of Faisalabad Medical University (FMU). Biopsy breast tissue samples were collected from patients who suffered from breast cancer. After collection biopsy samples were preserved in 10% formalin and 0.9% normal saline (NS) solution. Tissue samples were collected for RNA extraction and histopathology irrespectively.
Sample Processing for Expression analysis
Biopsy samples were further processed for mRNA isolation, performed manually by this standard protocol[18].cDNA was synthesized by the following standard protocol [19].qRT-PCR was performed followed by conventional PCR amplification of first strand cDNA[20]using specifically designed primers.
Histopathological Examination
For routine examination of histopathology, biopsy samples from breast cancer patients were collected in 10% formalin solution. Tissue specimens were embedded in paraffin before sectioning. Tissue was stained by using Haematoxylin and Eosin (H & E) stain for staining and analyzed under a microscope [21].
Statistical Analysis
Two-way ANOVA and DMR tests were used to examine graphical data statistically[22]. Graph pad prism 6 was used to draw graphs.
Results
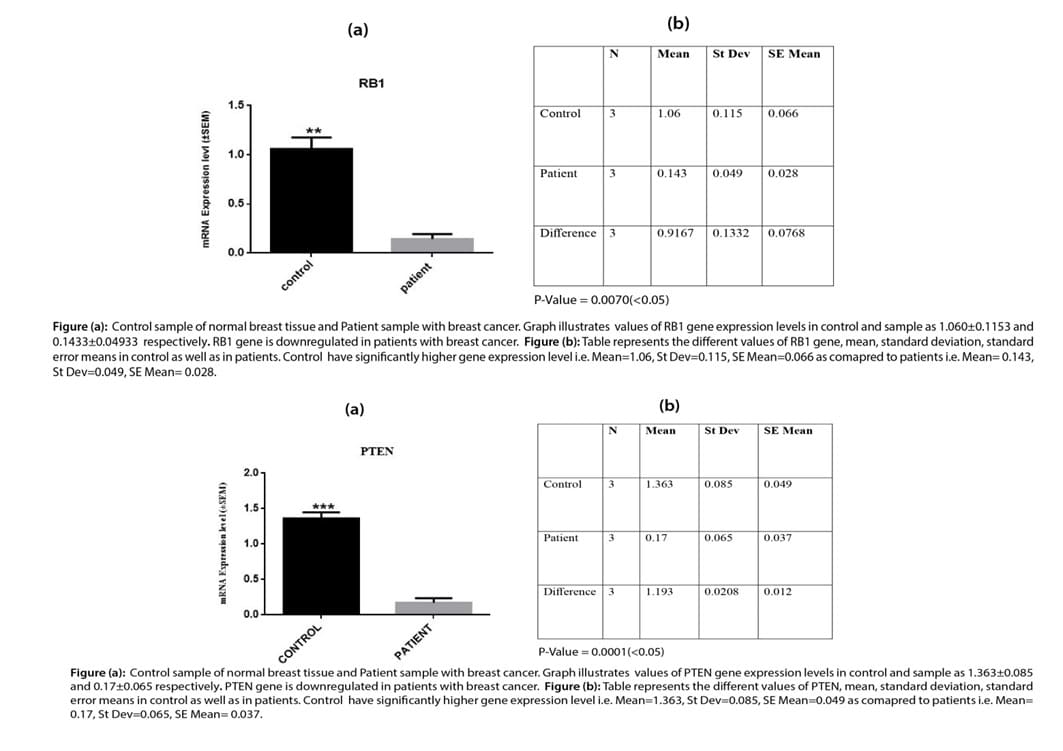
Relative gene expression T-Test for PTEN and RB1gene
The graph obtained from data analysis showed the relative gene expression of the PTEN gene and RB1 gene in control and patient samples as 1.363±0.085 and 0.17±0.065 for the PTEN gene respectively and 1.060±0.1153 and 0.1433±0.04933for RB1 gene respectively. Mean, standard deviation, and standard error mean were calculated using Two way ANOVA and DMR tests. Results showed that the PTEN and RB1 gene expression level is significantly high expression level in the control samples.
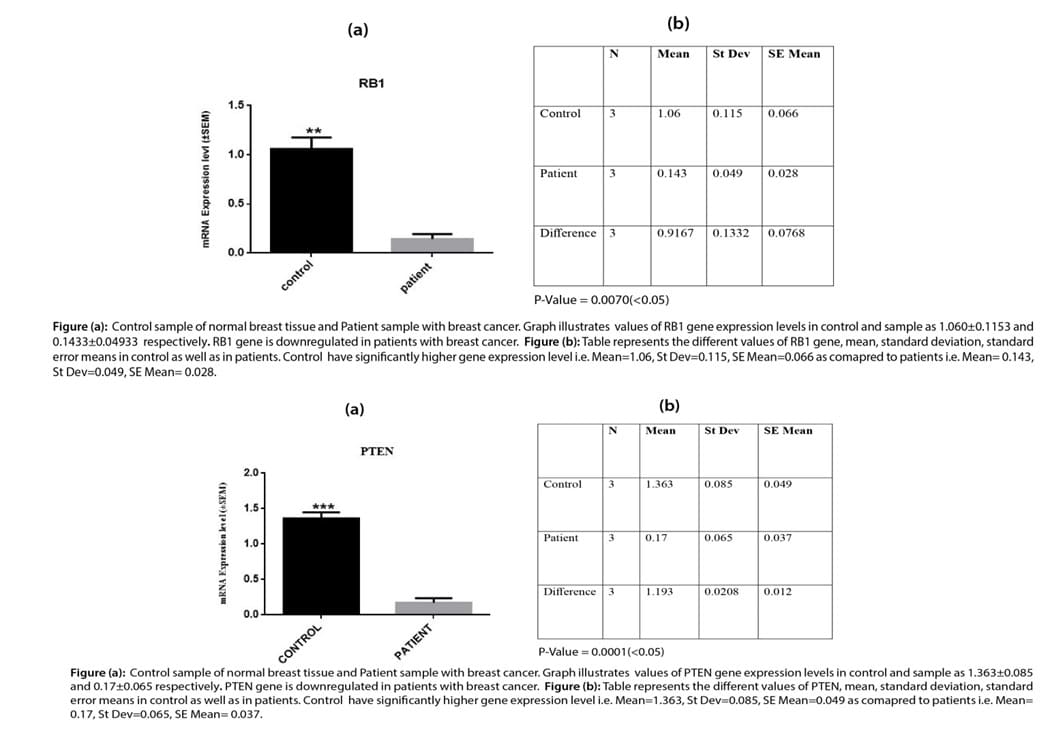
Relative gene expression T-Test forKRAS and Srcgene
The graph obtained from data analysis showed the relative gene expression of KRASgene and Src gene in control and patient samples as2.120±0.2352 and 3.377±0.25725 for KRAS gene respectively and1.283±0.1026 and 3.467±0.2857 forSrc1 gene. Mean, standard deviation, and standard error mean were calculated using two-way ANOVA and DMR tests. Results showed that KRAS and Src gene expression level is significantly high expression level in the patient’s samples.
Relative gene expression T-Test for MIR-140, MIR-145, MIR-238
The relative gene expression analysis of MIR-140, MIR-145, and MIR-238 genes showed an up-regulation in breast cancer patients. The values obtained from graphical analysis are 1.330±0.2107 (control) and 3.560±0.33 (patient) for MIR140, 1.263±0.08737 (control) and 3.510±0.4762 (patient) for MIR145, and 1.087±0.1422 (control) and 3.487±0.2577 (patient)for MIR238.Mean, standard deviation, and standard error mean was calculated using two-way ANOVA and DMR tests showed that MIR-140, MIR-145-, and MIR 238 gene expression level is significantly high expression level in patients’ samples.
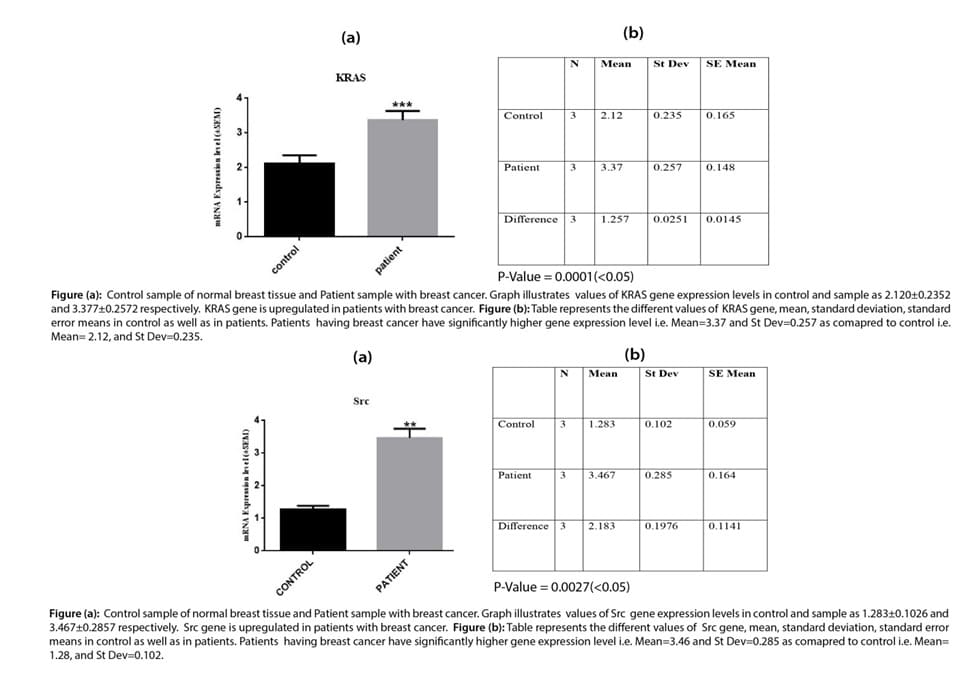
Histopathological examination of adenocarcinoma, ductal carcinoma, fibrous carcinoma, and lobular carcinoma in breast tissues
Histopathological examination of adenocarcinoma: Image A, shows normal breast tissue with the proper symmetrical shape of glandular tissue the lumen of the gland, lobules, lobular epithelium, and lobular ducts are present in normal histological confirmation while images B, C, and D indicates the breast tissue with cancerous growth of cells. The normal structure of glands, ducts, lobules, and the epithelium is lost and there are pycnotic nuclei and hypertrophy of epithelial cells is also evident in the lobular cuboidal epithelium.
Histopathological examination of ductal carcinoma: Image E is taken from normal breast tissues which indicates the normal functional histology of breast tissues with a single layer of simple cuboidal epithelium and myoepithelial cells with normal lobular and glandular tissue while images F, G, and H are taken from cancerous tissue of breast cancer affected patients where the structure of gland is completely lost with hypertrophy of glandular cell cuboidal epithelium and multilayering of glandular epithelial cells. In this patient ductal carcinoma was diagnosed because the ductal structure of the gland is destroyed and ducts of glands are blocked with the growth of fibrous connective tissue inside the lumen of ducts and as a result, there will be a complete loss of glandular secretion.
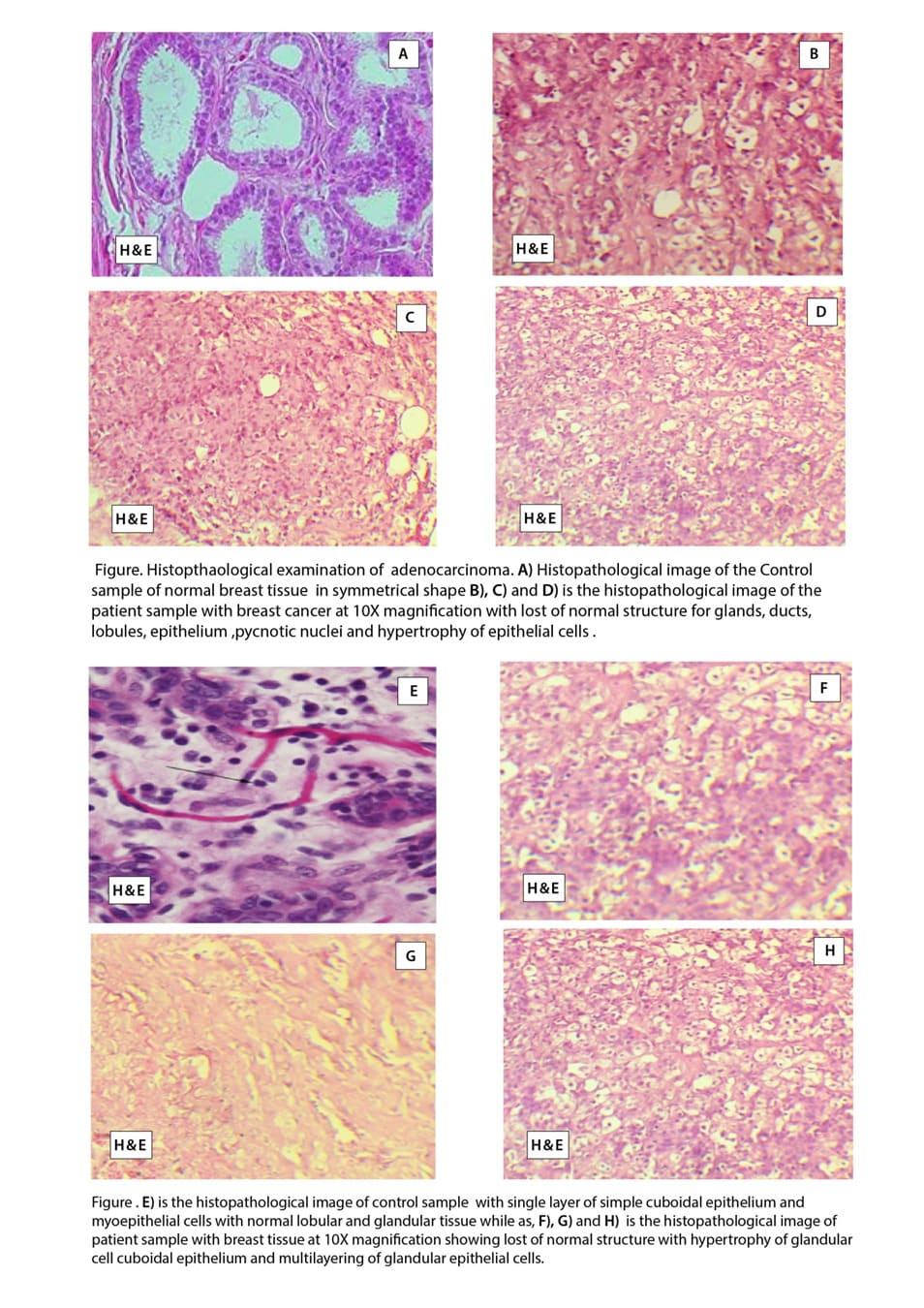
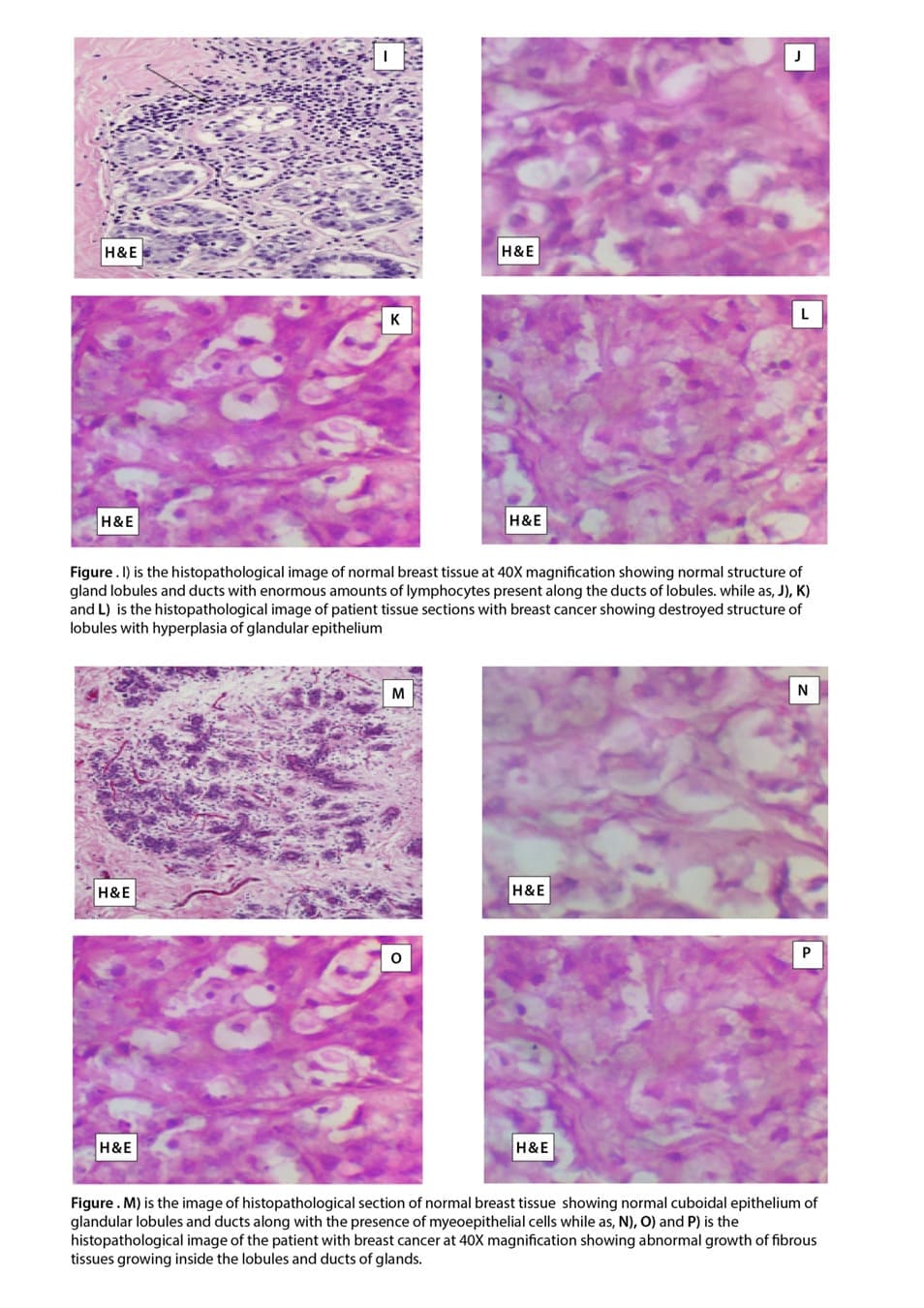
Histopathological Examination of lobular carcinoma: Image I, shows the histology of normal breast tissue in the parous female after menopause which indicates the normal structure of gland lobules and ducts with enormous amounts of lymphocytes present along the ducts lobules. Image J, K, and L is the histology of breast tissue from breast cancer patients which typically indicates lobular carcinoma in which the normal structure of lobules is destroyed and there is hyperplasia of glandular epithelium.
Histopathological examination of fibrous carcinoma: Image M, indicates the normal glandular structure of normal breast tissue with the normal cuboidal epithelium of glandular lobules and ducts along with the presence of myoepithelial cells. The lumen of ducts is normal where secretion of the gland is poured.
Images N, O, and P indicate the histology of fibrous carcinoma where the fibrous tissue growth is uncontrolled and fibrous tissue starts growing inside the lobules and ducts of glands.
Discussion
Breast cancer is the most commonly identified malignancy and the major contributor to cancer-associated deaths in females worldwide[23]. There is an upward trend in breast cancer burden in Pakistan, with one in every nine women having a lifetime chance of developing breast cancer [24]. The purpose of this study was to analyze the expression of tumor suppressor gene (PTEN and RB1), proto-oncogene (Src and KRAS), and micro RNA-140/145-238 in control and breast cancer patients along with the histopathological examination of adenocarcinoma, ductal carcinoma, fibrous carcinoma, and lobular carcinoma in breast tissues breast cancer patients.
Inside a normal cell, the microRNA-145 controls the growth and regulation of cells acting as a tumor suppressor and controlling the growth of normal cells but mutations and disorganized expressions can result in variable cancers. Previous studies report a significant increase in microRNA-140 expression levels in breast cancer patients with later stages[25]. The results of our study showed that there exists a direct relationship between the last breast cancer stage, metastasis, and the up-regulation of microRNA-140. In addition, a progressive increase in microRNA-140 was found in metastatic breast cancer compared to primary tumors[26]. Similarly, another study also reported elevated levels of miRNA-140 in cancers for promoting cell invasion [27,28].
The Preceding research on microRNA-145 revealed an important role played by this miRNA in the development and progression of breast carcinogenesis in human females. Our study reported that microRNA-145 is highly up-regulated in breast cancer patients. It is observed that the Rho factor which is encoded by the RTKN gene and inside the normal cell is present in low concentration but if miRNA-145 suppresses this RTKN gene it results in the origination of cancer cell lines. It is reported that over-expression of MicroRNA-145 results in resistance to apoptosis and the development of carcinogenesis [29].A mutational change in miRNA-145 results in the loss of its growth-promoting function resulting in the initiation and development of breast carcinogenesis[30]. A study reported the over-expression of micro RNA 145 in late stages of cancer growth i.e. stage III and stage IV similar to our results [31].
Similarly, miRNA-238 acts as a proto-oncogene in breast cancer-positive patients indicating its upregulation which results in carcinogenic growth in various tissues of the body including breast tissue. Our study showed that up-regulation of miRNA-328 is responsible for the progression of breast cancer in females which accept the previous findings.
Upregulation of the Src gene results in an increased level of expression of various growth factors including growth factors for epidermis, growth factors present on the surface of blood vessels
growth factors for fibroblasts may be due to increased expression of the Src gene in breast cancer [32]. Results of our study indicated the upregulation of the Src gene in breast cancer patients as compared to normal patients which indicate that the role of Src in a normal cell is as proto-oncogenic. A similar study depicted the up
regulation of Src in breast cancer while studying the importance of Src as a potential target for treatment [33].
KRAS plays a significant role in signaling for cellular proliferation and growth and mutations in KRAS have been directly associated with breast cancer. Our study showed high levels of KRAS expression in breast cancer patients as compared to control patients. A study reported a higher level of mRNA expression of the KRAS gene with a worse prognosis in breast cancer patients which relates to our findings[34]. A similar study was conducted to show the up-regulation of KRAS genes in variable cancers due to mutation[35].
It has been revealed that PTEN is concerned with many processes of cells such as cell growth [36] apoptosis[37], angiogenesis [38], migration, and invasion through interfering with various signalling pathways especially PIP3, PTEN/PI3K/
AKT signalling pathways are greatly involved in oncogenesis. PTEN is rarely mutated in NSCLC but the loss of its proteins is the most common event that takes place in breast cancer [39]. Our results showed that PTEN is a hereditary gene that is up-regulated in breast cancer and our findings accepts sthe role of PTEN from previous studies [40-43].
RB1 is a tumor suppressor gene and plays its role in controlling the cell cycle and apoptosis. Significant loss of RB1 leading to triple-negative breast cancer has been reported in previous studies due to mutation [44]. The results obtained for our study also shows the down-regulation of RBA1 genes in breast cancer patients which is also confirmed by the results of another study by Bondhopadhyay in 2021[45]. Similarly, frequent loss of the RB1 gene in metastatic cancers like breast and prostate cancers has also been confirmed by another study which agrees with our findings [46].
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
- Mahmood S, Rana TF, Ahmad M (2006) Common determinants of Ca breast-a case control study in Lahore. Annals of King Edward Medical University, 12.
- Arnold M, Soerjomataram I, Ferlay J, Forman D (2015) Global incidence of oesophageal cancer by histological subtype in 2012 Gut, 64: 381-387.
- Majeed Ai, Jadoon M, Riazuddin S, Akram J (2016) Awareness and screening of breast cancer among rural areas of Islamabad capital territory Pakistan. Annals of PIMS ISSN :1815, 2287.
- Hussain, Majeed A, Masood I, Ashraf W, Imran I, et al. (2022) A National Survey To Assess Breast Cancer Awareness Among The Female University Students Of Pakistan. Plos One, 17: E0262030.
- Saslow D, Boetes C, Burke W, Harms S, Leach M O, et al. (2007) American Cancer Society Guidelines For Breast Screening With Mri As An Adjunct To Mammography.Ca: A Cancer Journal For Clinicians, 57: 75-89.
- Foundation, N. B. C. 2022. Other Breast Cancer Genes.
- Pecorino L. (2021) Molecular Biology Of Cancer: Mechanisms, Targets, And Therapeutics, Oxford University Press.
- Papa A, Pandolfi P P (2019) ThePten–Pi3k Axis In Cancer. Biomolecules, 9, 153.
- Bhateja P, Chiu M, Wildey G, Lipka MB, Fu P, et al. (2019) Retinoblastoma Mutation Predicts Poor Outcomes In Advanced Non Small Cell Lung Cancer. Cancer Medicine, 8, 1459-1466.
- Wheeler DL, Iida M Dunn, E. F (2009) The Role Of Src In Solid Tumors. The Oncologist, 14, 667-678.
- Puls LN, Eadens M, Messersmith W (2011) Current Status OfSrc Inhibitors In Solid Tumor Malignancies. The Oncologist, 16, 566-578.
- Goitre L, Trapani E, Trabalzini L, Retta S. F (2014) TheRas Superfamily Of Small Gtpases: The Unlocked Secrets. RasSignaling, 1-18.
- Jančík S, Drábek J, Radzioch D, Hajdúch M (2010) Clinical Relevance Of Kras In Human Cancers. Journal Of Biomedicine And Biotechnology, 2010.
- Mcglynn LM, Kirkegaard T, Edwards J, Tovey S, Cameron D. (2009) Ras/Raf-1/Mapk Pathway Mediates Response To Tamoxifen But Not Chemotherapy In Breast Cancer Patients. Clinical Cancer Research, 15: 1487-1495
- Zhang Z, Wang Y, Vikis H.G, Johnson L, Liu G (2001) Wildtype Kras2 Can Inhibit Lung Carcinogenesis In Mice. Nature Genetics, 29: 25-33.
- Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A (2006) A Microrna Expression Signature Of Human Solid Tumors Defines Cancer Gene Targets. Proceedings Of The National Academy Of Sciences, 103: 2257-2261.
- Reddy, K. B (2015) Microrna (Mirna) In Cancer. Cancer Cell International, 15, 1-6.
- Sorlie T, Tibshirani R, Parker J, Hastie T,Marron J. S (2003) Repeated Observation Of Breast Tumor Subtypes In Independent Gene Expression Data Sets. Proc Natl AcadSci U S A, 100: 8418-23.
- Wiame I, Remy S, Swennen R, Sági L (2000) Irreversible Heat Inactivation OfDnase I Without Rna Degradation. Biotechniques, 29, 252-256.
- Bustin S (2002) Invited Review Quantification Of Mrna Using Real-Time Reverse Transcription Pcr (Rt-Pcr): Trends And Problems. Journal Of Molecular Endocrinology, 29: 23-39.
- Kanel GC, Korula J, Renshaw A (2005) Atlas Of Liver Pathology 2nd Edition Lww.
- Masri BA, Duncan C. P, Beauchamp C P, Paris N J, Arntorp J (1995) Effect Of Varying Surface Patterns On Antibiotic Elution From Antibiotic-Loaded Bone Cement. The Journal Of Arthroplasty 10:453-459.
- Suleiman AK (2014) Awareness And Attitudes Regarding Breast Cancer And Breast Self-Examination Among Female Jordanian Students. Journal Of Basic And Clinical Pharmacy 5: 74.
- Tamura M, Gu J, Matsumoto K, Aota S-I, Parsons R, Yamada KM (1998) Inhibition Of Cell Migration, Spreading, And Focal Adhesions By Tumor Suppressor Pten. Science 280: 1614-1617.
- Zaheer S, Shah N, Maqbool SA, Soomro NM (2019) Estimates Of Past And Future Time Trends In Age-Specific Breast Cancer Incidence Among Women In Karachi, Pakistan 2004–2025. BMC Public Health 19: 1-9.
- Zhai Z, Qu X, Li H, Ouyang Z, Yan W, et al. (2015) Inhibition Of Mda-Mb-231 Breast Cancer Cell Migration And Invasion Activity By Andrographolide Via Suppression Of Nuclear Factor-Κb-Dependent Matrix Metalloproteinase-9 Expression Corrigendum In/10.3892/Mmr. 2019.9833. Molecular Medicine Reports, 11, 1139-1145.
- Yang S, Li Y, Gao J, Zhang T, Li S, et al. (2013) Microrna-34 Suppresses Breast Cancer Invasion And Metastasis By Directly Targeting Fra-1. Oncogene 32: 4294-4303.
- Zheng M, Liu J, Meng C, Tang K, Liao J (2021) Prognostic AndClinicopathological Importance Of Microrna-140 Expression In Cancer Patients: A Meta-Analysis. World Journal Of Surgical Oncology 19: 1-9.
- Hannafon BN, Ding WQ (2019) Functional Role OfMirnas In The Progression Of Breast Ductal Carcinoma In Situ. The American Journal of Pathology 189: 966-974.
- Communications Biology. 3: 1-12.
- Lotterman CD, Kent OA, Mendell JT (2008) Functional Integration OfMicrornas Into Oncogenic And Tumor Suppressor Pathways. Cell Cycle 7: 2493-2499.
- Jiang X, Zhou Y, Sun AJ, Xue JL (2018) Neat1 Contributes To Breast Cancer Progression Through Modulating Mir‐448 And Zeb1. Journal Of Cellular Physiology 233: 8558-8566.
- Manvati S, Mangalhara KC, Kalaiarasan P, Chopra R, Agarwal G et al. (2019). Mir-145 Supports Cancer Cell Survival And Shows Association With Ddr Genes, Methylation Pattern, And Epithelial To Mesenchymal Transition. Cancer Cell International 19: 1-12.
- Bild A H, Yao G, Chang JT, Wang Q, Potti A, et al. (2006) Oncogenic Pathway Signatures In Human Cancers As A Guide To Targeted Therapies. Nature 439: 353-357.
- Belli S, Esposito D, Servetto A, Pesapane A, Formisano L, et al. (2020) C-Src And Egfr Inhibition In Molecular Cancer Therapy: What Else Can We Improve? Cancers 12: 1489.
- Hwang KT, Kim BH, Oh S, Park SY, Jung J, et al. (2019) Prognostic Role of KrasMrna Expression In Breast Cancer. Journal of Breast Cancer 22: 548-561.
- Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, et al. (2020) KrasSignaling Enriched Triple Negative Breast Cancer Is Associated With FavorableTumor Immune Microenvironment And Better Survival. American Journal of Cancer Research, 10: 897.
- Stewart AL, Mhashilkar AM, Yang XH, Ekmekcioglu S, Saito Y, et al. (2002) Pi3k Blockade By Ad-Pten Inhibits Invasion And Induces Apoptosis In Radial Growth Phase And Metastatic Melanoma Cells. Molecular Medicine, 8: 451-461.
- Dothager RS, Putt KS, Allen BJ, Leslie BJ, Nesterenko V, et al. (2005) Synthesis And Identification Of Small Molecules That Potently Induce Apoptosis In Melanoma Cells Through G1 Cell Cycle Arrest. Journal of The American Chemical Society, 127: 8686-8696.
- Castellino RC, Durden DL. (2007) Mechanisms Of Disease: The Pi3k–Akt–PtenSignaling Node—An Intercept Point For The Control Of Angiogenesis In Brain Tumors. Nature Clinical Practice Neurology, 3: 682-693.
- Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S et al. (1998) Mutation Analysis Of The Pten/Mmac1 Gene In Lung Cancer. Oncogene, 17: 1557-1565.
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490: 61-70.
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, et al. (2015) Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer Cell, 163: 506-519.Mendes-Pereira A M,SimsD,Dexter T, Fenwick K, Assiotis I. Kozarewa I et al. 2012 Genome-Wide Functional Screen Identifies A Compendium Of Genes Affecting Sensitivity To Tamoxifen. Proceedings of The National Academy Of Sciences, 109: 2730-2735.
- Salmani H, Hosseini, zarnezhad A, Ahmad H (2018) Pten And P53 Gene Expressions In Breast Cancer Specimens And Their Clinicopathological Significance. Middle East Journal of Cancer, 9: 105-111.
- Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, et al. (2016) Rb1 Deficiency In Triple-Negative Breast Cancer Induces Mitochondrial Protein Translation. The Journal of Clinical Investigation, 126: 3739-3757.
- Bondhopadhyay B, Sisodiya S, Kasherwal V, Nazir SU, Khan A, et al. (2021) The Differential Expression of PromyelocyticLeukemia (Pml) And Retinoblastoma (Rb1) Genes In Breast Cancer. Meta Gene, 28: 100852.
- Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, et al. (2020) Pan-Cancer Molecular Analysis of theRbTumor Suppressor Pathway.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.