Programmed response evaluation and prediction of GBMs using Artificial Intelligence (AI) and Imaging with the description of a swarmdeepsurv model which innovatively combines SI (Swarm Intelligence) - based feature selection and deepsurv: a brief overview
by Diana Donatello*
MD,Radiologist,Independent Researcher, Costa Contina Street n.19, 66054, Vasto, Chieti, Italy
*Corresponding author: Diana Donatello, MD,Radiologist, Independent Researcher, Costa Contina Street n.19, 66054, Vasto, Chieti, Italy
Received Date: 07 September 2024
Accepted Date: 16 September 2024
Published Date: 18 September 2024
Citation: Donatello D (2024) Programmed response evaluation and prediction of GBMs using Artificial Intelligence (AI) and Imaging with the description of a swarmdeepsurv model which innovatively combines SI (Swarm Intelligence) - based feature selection and deepsurv: a brief overview. J Surg 9: 11140 https://doi.org/10.29011/2575-9760.11140
Abstract
Object:To describe the main novel tools of AI in brain imaging.
Methods: review of the literature.
Description: This review describes the development of novel analysis tools,using state-of-the-art AI that can make sense of the data acquired during GBM diagnosis and treatment in an effort to help improve and standardize the patient pathway for this disease [1-4].
Conclusion: Nowadays are being born new revolutionary programs for theprediction and survival of GMBs based on AI.
Object:
The Generation of new data flow channels for integrating imaging and patient demographic data in GBMs and combine research into a software prototipe for use within future clinical trials and further clinical implemantion is described both in the study of Waqar et al and in the study of Molina et al. [5,6] The identifcation of new biomarkers of GBMs response prediction and tumour grading using AI is best described in the study of Vieira et al. [7] Finally the study based on a prototipe of SwarmDeepSurvival model for use prognostic models is analized by Al-Tashi et al. [8].
Description:
Image-Based Tumour Grading
MRI is typically performed prior to surgery in GBMs diagnosis. The evolving field of radiomics is utilized to extract high-order features from MRI images (e.g. tumour shape and texture), and determine image-based models of tumour grade. Recorded GBMs grades identified using histopathology (in case of surgery and after surgery) will be used as “gold-standard” in this target [2,3,5,6,9].
Programmed Detection of Heterogeneus GBM Environments, Through Imaging
Conventional radiological measures of tumour size using CT or MRI fail as response biomarkers due to the inter-tumoural heterogeneity encountered in GBMs.
There is evidence that AI can be used to detect heterogeneous subcompartments from multi-parametric MRI in response to radio/ chemoteraphy, for example: multi-parametric MRI images could be combined using to categorize regions into one of three possible tissue groups:
1) Cellular vascularized tumour
2) Poorly vascularized tissue
3) Dead tissue
This classification scheme is also applied following radio/chemiotherapy to assess for changes in the volume of each compartment. A heterogeneous pattern colud be identified before treatment, with the majority of the tumour consisting of highly vascular regions. Following radio/chemiotherapy, the proportion of poorly vascularised tissue could increase indicating a disruption in the blood supply to the tumour (Figure1)[6]. Using the collective data archive, we can study the biological basis of these regions, and develop response/ prognostic models that combine imaging phenotypes with regional histopathology and molecular profiling of tissues [4].
Clinical utility of habitat imaging in glioblastoma: assessment of changes pre and post-radiotherapy. These figures demonstrate the clinical utility of habitat imaging in glioblastoma pre and post-radiotherapy
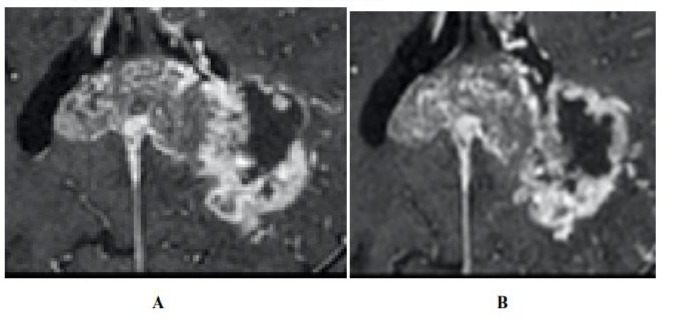
Figure 1:T1 with contrast demonstrates no significant changes in tumour anatomy pre treatment(A) and post treatment (B). [6]
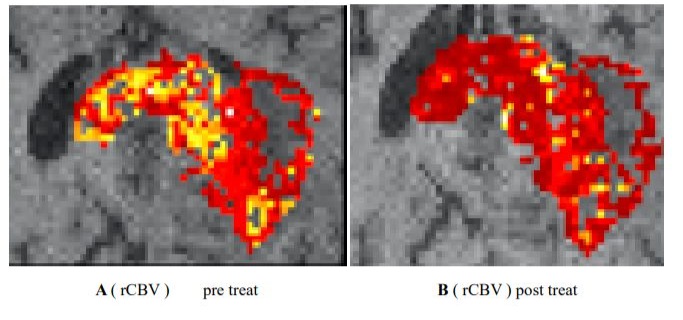
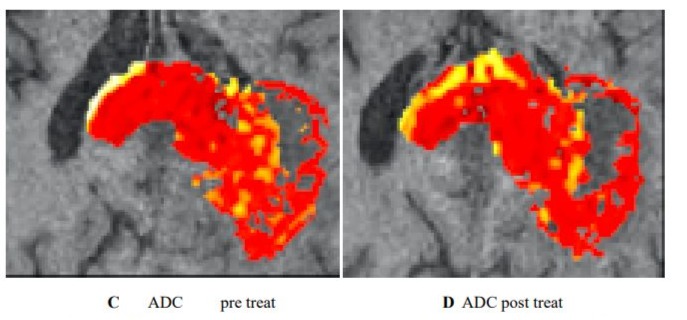
Figure 2 :diffusion and perfusion MRI scans demonstrate changes in tumour physiology with treatment with a decrease in rCBV pre treatment (A) Imaging habitats map where each voxel is labelled according to both rCBV and ADC values(C). After radiotherapy, the increase was in a habitat defined by low rCBV and low ADC (B-D). [6]
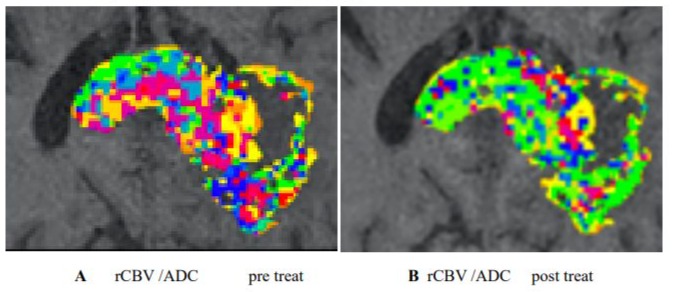
Figure 3. The maximum ecrease was in a habitat defined by high rCBV and medium ADC (A,B) . Habitats that are more resistant to treatment can be spatially visualised and offered targeted therapy. [5,6].
Multi-Omic Prediction Biomarkers in GBM
Investigators have demonstrated utility for predictive models of patient prognosis based on patient demographic information and GBMs descriptors such as size and location. These descriptors cannot account for spatial tumour heterogeneity in the way imaging can. So a new AI toolkit using deep-learning that can combine imaging data with patient demographic information to determine predictive models of local recurrence, treatmente toxicity and patient survival can be potentially create [10,11,7].
Develop a Survival Model, Swarmdeepsurv, which Innovatively Combines SI-Based Feature Selection and Deepsurv.
Integrate swarm intelligence with deep survival modeling to create a new platform termed SwarmDeepSurv to discover prognostic models from high-dimensional features. Nowadays we can develop an innovative survival prediction framework known as SwarmDeepSurv that can predict survival in different cancer types using a large multicenter radiomics dataset, the network developed by Al-Tashi Q at al. Figure 4 [8,12].
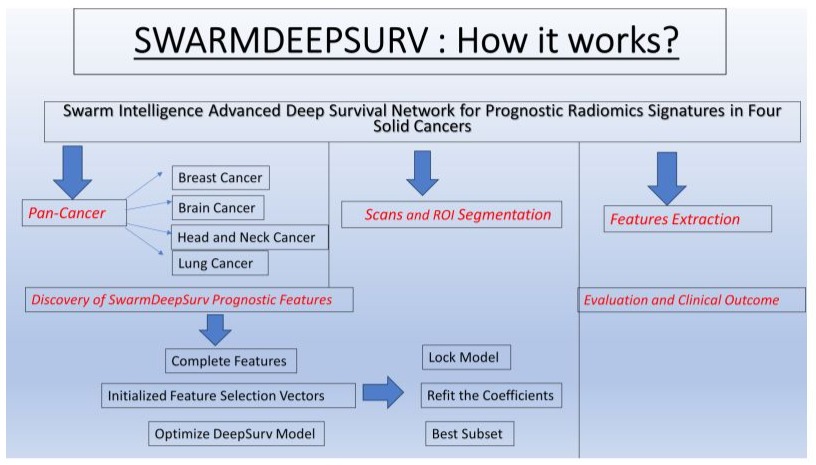
Figure 4: Graphic Abstract.
Conclusion:
New AI methodologies for combinating imaging data with tumor grading and demographic data within GBMs are growing. The field of radiomic features extraction channels for GBMs is in expansion. With this minireview study we can undestand the potential of multi-omic, predictive biomarkers of GBMs response to chemotherapy and validation of biomarkers using available histopathology data. Finally the development of a the platform called SwarmDeepSurv can predict survival outcomes.
References
- Gonzales A, Guruswamy G, Smith SR (2023) Synthetic data in health care: A narrative review. PLOS Digit Health 2: e0000082.
- al-Rifaie MM, Aber A, Hemanth DJ (2015) Deploying swarm intelligence in medical imaging identifying metastasis, micro-calcifications and brain image segmentation. IET Syst Biol 9: 234-244.
- Liu Y, Xu X, Yin L, Zhang X, Li L, et al. (2017) Relationship between Glioblastoma Heterogeneity and Survival Time: An MR Imaging Texture Analysis. AJNR Am J Neuroadiol 38: 1695-1701.
- Frederico SC, Darling C, Bielanin JP, Dubinsky AC, Zhang X, (2023) Neoadjuvant immune checkpoint inhibition in the management of glioblastoma: Exploring a new frontier. Front Immunol 14: 1057567.
- Molina D, Pérez-Beteta J, Luque B, Arregui E, Calvo M, et al. (2016) Tumour heterogeneity in glioblastoma assessed by MRI texture analysis: a potential marker of survival. Br J Radiol 89: 20160242.
- Waqar M, Van Houdt PJ, Hessen E, Li KL, Zhu X, et al. (2022) Visualising spatial heterogeneity in glioblastoma using imaging habitats. Front Oncol 12: 1037896.
- Vieira FG, Bispo R, Lopes MB. Integration of Multi-Omics Data for the Classification of Glioma Types and Identification of Novel Biomarkers. Bioinform Biol Insights 18: 11779322241249563.
- Al-Tashi Q, Saad MB, Sheshadri A, Wu CC, Chang JY, et al. (2023) SwarmDeepSurv: swarm intelligence advances deep survival network for prognostic radiomics signatures in four solid cancers. Patterns (N Y) 4: F100777.
- Ronvaux L, Riva M, Coosemans A, Herzog M, Rommelaere G, et al. (2022) Liquid Biopsy in Glioblastoma. Cancers (Basel) 14: 3394.
- arzegar Behrooz A, Latifi-Navid H, da Silva Rosa SC, Swiat M, Wiechec E, et al. (2023) Integrating Multi-Omics Analysis for Enhanced Diagnosis and Treatment of Glioblastoma: A Comprehensive DataDriven Approach. Cancers (Basel) 15: 3158.
- Arslan S, Schmidt J, Bass C, Mehrotra D, Geraldes A, et al. (2024) A systematic pan-cancer study on deep learning-based prediction of multi-omic biomarkers from routine pathology images. Commun Med (Lond) 4: 48.
- Majumder S, Katz S, Kontos D, Roshkovan L (2024) State of the art: radiomics and radiomics-related artificial intelligence on the road to clinical translation. BJR|Open 6: tzad004.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.