Comprehensive Pan-cancer Analysis Identified ZCCHC3 as an Immunological and Prognostic Biomarker
by Xuehan Gao1# Pinzhi Dong1#, Linna Wei1, Jin Chen1, Haiyan Wang2, Ming Qin1*, Junmin Luo1*, Jihong Feng3*
1Department of Immunology, Zunyi Medical University, Zunyi 563000, China
2School of Public Health, Zunyi Medical University, Zunyi 563000, China
3Department of oncology, Lishui People’s Hospital, Sixth Affiliated Hospital of Wenzhou Medical University, Lishui 323000, China # XHG and PZD contributed equally to this work.
*Corresponding authors: Ming Qin, Department of immunology, Zunyi Medical University, GuiZhou Province, 563000 P.R.China.
Junmin Luo, Department of immunology, Zunyi Medical University, GuiZhou Province, 563000 P.R.China.
Jihong Feng, Department of oncology, Lishui People’s Hospital, Zhejiang Province, 323000 P.R.China.
Received Date: 29 August, 2023
Accepted Date: 08 September, 2023
Published Date: 11 September, 2023
Citation: Gao X, Dong P, Wei L, Chen J, Wang H, et al. (2023) Comprehensive Pan-cancer Analysis Identified ZCCHC3 as an Immunological and Prognostic Biomarker. Ann med clin Oncol 5: 154. https://doi.org/10.29011/2833-3497.000154
Abstract
Purpose: ZCCHC3 may be closely associated with the development of tumors. However, the detailed function and role of ZCCHC3 in pan-cancer are largely unknown and require further in-depth investigation. Consequently, this study aims to investigate the biological functions of ZCCHC3, and its potential to predict prognosis and immunotherapy response in pancancer. Methods: We applied multiple public databases such as TCGA, TIMER, GTEx, CCLE, and HPA to explore ZCCHC3 expression in tumors. Univariate Cox regression analysis were used to detect the effects of ZCCHC3 on OS, DSS, DFI, and PFI in these patients. Subsequently, the correlation coefficient between ZCCHC3 levels and immune infiltration in different cancer types was used using TIMER2.0. Finally, the relationship between ZCCHC3 and tumor immune regulatory genes, immune checkpoint, TMB, and MSI were investigated. Results: ZCCHC3 expression was elevated in most tumor tissues compared to normal tissues, and was positively or negatively correlated with the prognosis of different tumors, positively correlated with immune cell infiltration and immune checkpoint gene expression in various tumors. Conclusion:Comprehensive pan-cancer analysis identified ZCCHC3 as an immunological and prognostic biomarker.
Keywords: ZCCHC3; Prognosis; Immune analysis; Pan- cancer; The Cancer Genome Atlas
Introduction
Cancer development is a complex process that involves the participation of many signaling pathways and genes [1-2]. Pan-cancer analysis refers to the cross-sectional comparison of certain features across multiple tumour types through the use of bioinformatics analysis tools, with the implication of applying diagnosis and treatment to a wider range of tumours through cross-tumour similarity [3]. In recent years, with the rapid development of sequencing technologies and the establishment of online databases, a large influx of data has provided the basis for comprehensive pan-cancer analysis to guide the diagnosis and treatment of tumours. Consequently, the identification of key genes between different cancer types can help in the diagnosis and treatment of cancer.
Zinc finger proteins are a series of proteins with a fingerlike shape, which have a relatively short sequence and need to be presented in a more stable condition by binding to zinc ions, and contain an 18-residue structural domain with the sequence CX2CX4HX4C [4]. A variety of zinc finger-containing eukaryotic proteins are involved in many aspects of nucleic acid metabolism, ranging from DNA transcription to RNA degradation, posttranscriptional gene silencing, and the biogenesis of small RNAs, which, in turn, regulate gene expression [4]. Zinc finger CCHCtype (ZCCHC) superfamily proteins are thought to bind with high affinity to single-stranded nucleic acids. In humans, 25 ZCCHC proteins are annotated in the HGNC database, and most members of the ZCCHC family of hyperproteins are involved in multiple steps of RNA transcription, biogenesis, splicing, and translation and degradation. Thus, zinc finger proteins are an important class of proteins that regulate gene expression and play important roles in life activities.
The CCHC-type zinc-finger protein ZCCHC3 is a CCHCtype zinc-finger protein, which was recently discovered to involve in antiviral innate immune responses [5]. Lian et al. [6]. revealed ZCCHC3 to be a co-sensor of cyclic GMP-AMP (cGAMP) synthase (cGAS) for the recognition of cytosolic dsDNA; cGAS catalyzes synthesis of the second messenger molecule cGAMP, which in turn binds and activates the adaptor STING to initiate an innate antiviral response. Furthermore, ZCCHC3 was shown to bind dsRNA and act as a positive regulator of RIG-I-like receptor (RLR), including RIG-I (retinoic acid-inducible gene-I) and MDA5, and Toll-like receptor 3 (TLR3) signaling. In addition, ZCCHC3 has been shown to be part of the SARS-CoV-2 virus protein interactome and to inhibit avian H9N2 virus and pseudorabies virus through type I IFN signaling. Briefly, ZCCHC3 promotes interactions between viral nucleic acids and pattern recognition receptors, including cGAS, RIG-I-like receptor and Toll-like receptor 3, thereby positively regulating RNA and DNA virus-triggered IFN signalling, suggesting a multifunctional role in antiviral innate immunity [7]. Notably, ZCCHC3 may play an important role in tumour development. Wang et al. found that FOXD3 Antisense RNA 1 (FOXD3-AS1) sponges miR-296-5p to elevate ZCCHC3 to facilitate malignancy in osteosarcoma, this may provide potential guidance for finding effective targets for the treatment of osteosarcoma [8]. Copy Number Alterations (CNAs) represent the most common genetic alterations identified in ovarian cancer cells, being responsible for the extensive genomic instability observed in this cancer. Marco et al. [9]. have identified 201 altered chromosomal bands and 3,300 altered genes in human ovarian cancer samples. Then, the 3,300 genes subjected to CNA identified here were compared to those present in the TCGA dataset. The analysis allowed the identification of 11 genes with increased CN and mRNA expression, Interestingly, ZCCHC3 is among the highly expressed genes. The above findings suggest that ZCCHC3 may have broad and diverse regulatory roles in cancer. Up to now, no studies have performed pan-cancer analyses of ZCCHC3. Therefore, we aimed to elucidate the role of ZCCHC3 in tumour immunomodulation and immunotherapy through a comprehensive pan-cancer analysis.
Materials and methods
ZCCHC3 Expression Pattern in Human Pan-Cancer
TCGA, a cornerstone of the cancer genomics projects, had characterized more than 20,000 primary cancer samples and corresponding non‐carcinoma samples from 33 types of cancers. In the present study, the TCGA‐processed level 3 RNA‐sequencing data sets, along with the corresponding clinical annotations, were obtained using the University of California Santa Cruz (UCSC) cancer genome browser (https:// tcga.xenahubs.net, accessed April 2020). The CCLE public project has comprehensively characterized a tremendous number of human tumour models both genetically and pharmacologically (https://portals.broadinstitute. org/ccle). To examine differential gene expression in cancers on a larger scale, the CCLE database, which contains RNA‐sequencing data sets for over 1,000 cell lines, was used. RNA sequencing data and clinical follow-up information for patients with 33 types of cancers.
Protein level analysis
The Human Protein Atlas (HPA: https://www.proteinatlas. org/) database was used to explore the protein level of ZCCHC3 in human tumor and normal tissues. GeneCards (https://www. genecards.org/) was used to visualise the subcellular locations of ZCCHC3. String (https://string-db.org/) database was used to construct the protein-protein interaction network (PPI) of ZCCHC3.
Pathological staging analysis
The expression data of ZCCHC3 gene in each sample were extracted from TCGA database, and the samples from Solid Tissue Normal, primary blood derived cancer-peripheral blood and Primary Tumor were further screened. Log2 (x+0.001) transformation was performed on each expression value, and finally, the cancer species with less than 3 samples in a single cancer species were eliminated, and finally the expression data of 26 cancer species were obtained. The expression difference between normal samples and tumor samples in each tumor was calculated by R software (version 3.6.4), and the difference significance was analyzed by unpaired Wilcoxon rank sum and signed rank tests.
Prognostic Analysis
The connection between the ZCCHC3 expression and the prognosis of patients, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) in 33 types of cancer was examined using forest plots.
ZCCHC3 and tumor Immune infiltration
We used the “Immune-Gene” module of the TIMER2.0 datebase to explore the association between ZCCHC3 expression and immune infiltrates across all TCGA tumors. The immune cells of Tregs, cancer-associated fibroblast, DC, Macrophage, and T cell CD8+ were selected.
Correlation analysis
We downloaded the uniformly normalized pan-cancer dataset: TCGA TARGET GTEx (PANCAN, N=19,131, G=60,499) from the UCSC (https://xenabrowser.net/) database, from which we further extracted the ZCCHC3 gene and 60 genes of two types of immune checkpoint pathways (Inhibitory, Stimulatory, screened samples from: Primary Solid Tumor, Primary Tumor, Primary Blood Derived Cancer - Bone Marrow, Primary Blood Derived Cancer-Peripheral Blood, we also filtered all normal samples and furthermore log2(x+0.001) transformed each expression value, next we calculated the spearman correlation between ZCCHC3 and marker genes of the five immune pathways. Tumor-Immune System Interaction DataBase (TISIDB) (http://cis.hku.hk/ TISIDB/) was utilized to explore the relationships of ZCCHC3 and immune modulators in pan-cancer.
Analysis of single-cell sequencing results from the TISCH database
The tumor immune single-cell Hub (TISCH) (http://tisch. comp-genomics. org/documentation/) is a scRNA-seq database that integrates the single-cell transcriptome profiles of nearly 2 million cells from 76 high-quality tumour datasets for 27 cancers. Single-cell sequencing aims to characterize the similarities and differences between different tumors or within the same tumor at the cellular level. Characterising the tumour microenvironment at single-cell resolution.
The expression levels of ZCCHC3 in immunotherapy patients were compared in TISMO database
The TISMO database (http://tismo.cistrome.org/) contains a large number of homologous mouse model data, including RNAseq data from 605 extracontogenetic samples of 49 homologous cancer cell lines from 23 cancers, 195 of which received cytoplasmic therapy; In addition, we included RNA-seq data from 1,518 in-person samples from 68 homologous mouse models of 19 cancers, 832 of which were from the immunecheckpoint blocking (ICB) study.
TIDE database predicts immunotherapy response
Tumor Immune Dysfunction and Exclusion (TIDE) database (http://tide. dfci. harvard. edu) integrating large-scale omics data and biomarkers from published ICB trials, non-immunotherapy tumor profiles, and CRISPR screenings. Select the “Biomarker Evaluation” module to compare ZCCHC3 with other published biomarkers. The area undercurve (AUC) of the receiver operating characteristic (ROC) was used to evaluate the predictive performance of the biomarker to the ICB response state.
Pan-Cancer Analysis of the Relationship between the ZCCHC3 Expression and TMB or MSI
The TMB and MSI scores were obtained from TCGA. Correlation analysis between the ZCCHC3 expression and TMB or MSI was performed using Spearman’s method. The horizontal axis in the figure represents the correlation coefficient between ZCCHC3 and TMB or MSI, the ordinate is different types of cancer, the size of the dots in the figure represents the size of the correlation coefficient, and the different colors represent the significance of the P value.
Results
ZCCHC3 expression Analysis in Pan-Cancer
Firstly, we evaluated the expression of ZCCHC3 in TCGA. The results showed that ZCCHC3 was highly expressed in COAD, COADREAD, ESCA, STES, STAD, HNSC, KIRC, LIHC, READ, PCPG, and CHOL, Down-regulated in 4 kinds of tumors, such as LUAD, PRAD, UCEC, and THCA (Figure 1A). On this basis, combining TCGA and GTEx databases revealed that ZCCHC3 was highly expressed in GBM, GBMLGG, LGG, BRCA, LUAD, ESCA, STES, KIPAN, COAD, COADREAD, PRAD, STAD, HNSC, KIRC, LUSC, LIHC, WT, SKCM, BLCA, THCA, READ, OV, PAAD, UCS, ALL, and CHOL 29 tumors, but low in UCEC (Figure 1B). The results of CCLE analysis showed that ZCCHC3 showed inconsistent gene expression levels in various cancer cell lines, SCLC, DLBC, NB, ALL showed relatively high gene expression (Figure 1C). Finally, the TIMER database was used to evaluate the expression of ZCCHC3 in pan-cancer. The results showed that the expression of ZCCHC3 in CHOL, COAD, HNSC, KIRC, LIHC, LUAD, LUSC, PCPG, PRAD, SKCM, and STAD was significantly higher than that in normal tissues (Figure 1D).
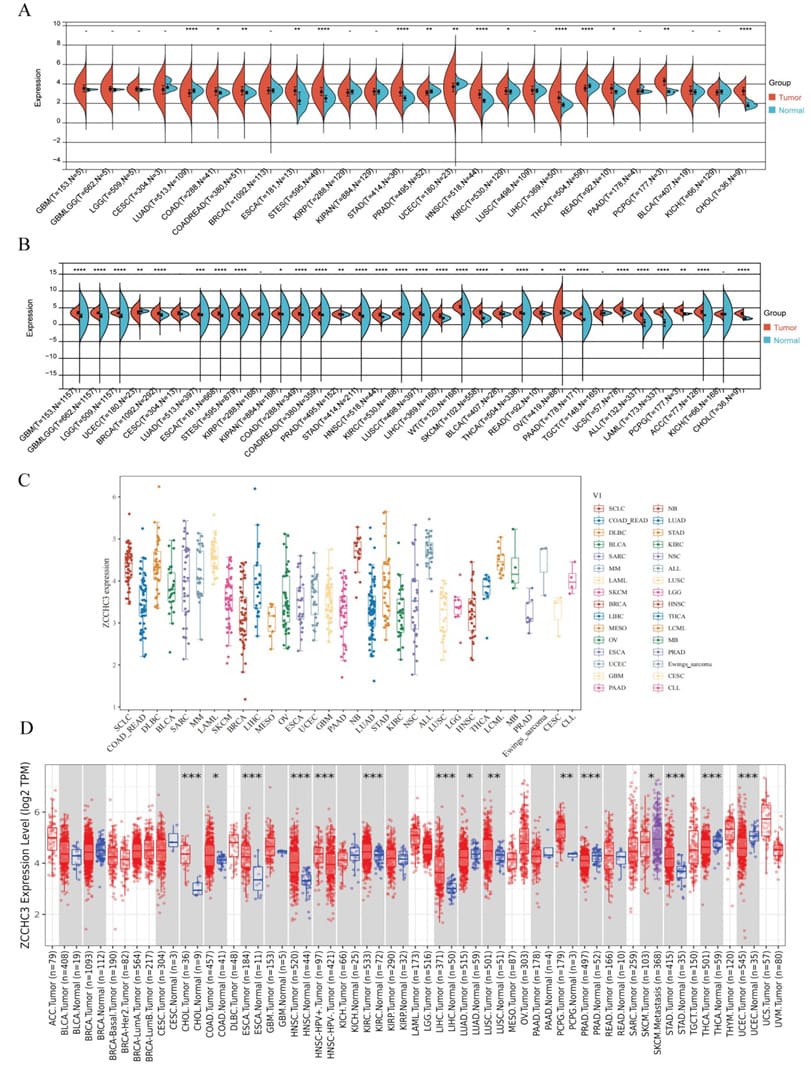
Figure 1: Pan-cancer ZCCHC3 expression (A) ZCCHC3 expression in tumor tissues from TCGA database; (B) Pan-cancer expression of ZCCHC3 between tumor tissues from TCGA database and normal tissues from TCGA and GTEx database; (C) mRNA expression levels of ZCCHC3 in various tumor cell lines from CCLE database; (D) ZCCHC3 expression in tumor tissues from TIMER database. The red and blue boxes represent tumor tissues and normal tissues, respectively.*p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001; ns, not significant.
To assess gene expression levels for all tumor stages, we compared ZCCHC3 expression in patients with stage I, II, III, or IV tumors. Significantly different in LUAD, COAD, COADREAD, KIPAN, KIRC, THYM, PAAD, and KICH (Figure 2A-H).

Figure 2: Pan-cancer ZCCHC3 expression in different WHO stages;(A) Pan-cancer differential expression of ZCCHC3 in WHO stages in indicated tumor types from TCGA database. *p < 0.05; **p < 0.01; ***p < 0.001, and ****p < 0.0001; ns, not significant; (B) LUAD; (C) COAD; (D) COADREAD; (E) KIPAN; (F) KIRC; (G) THYM; (H) PAAD; (I) KICH.
Protein Level of ZCCHC3
Based on datasets of the HPA, GTEx, and FANTOM5 (function annotation of the mammalian genome), we found that ZCCHC3 expression has a low tissue specificity, but enriched in Thymus and Placenta (Figure 1A; Supplementary Figure S1). Moreover, based on single-cell RNA-seq, we found that ZCCHC3 has low cell type specificity (Supplementary Figure S1). Furthermore, we explored the subcellular location of ZCCHC3 using the GeneCards database and showed that ZCCHC3 protein is mainly at the nucleus (Figure 3B). The protein interacting with ZCCHC3 was predicted by STRING, the results show that the interaction with ZCCHC3 protein mainly include DDX58, TRIM25, MOV10, IFIH1, TRIM56, CGAS, PABPC1, PAIR1, UPF1, SERBP1 (Figure 3C).
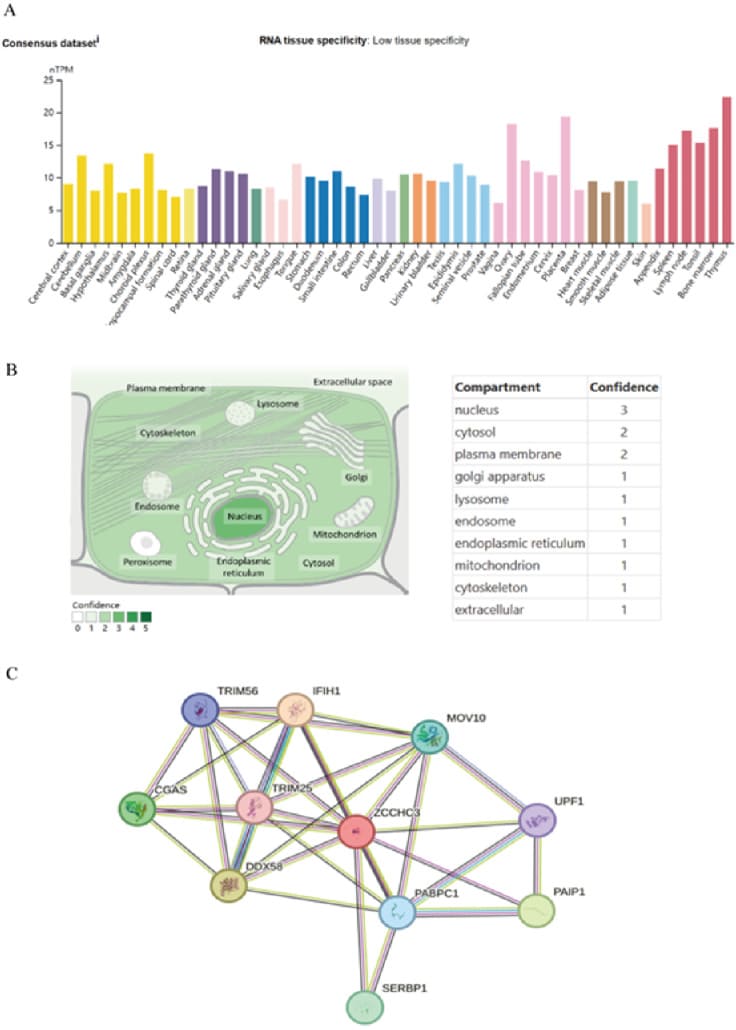
Figure 3: ZCCHC3 expression status in different tumors and normal tissues; (A) Consensus ZCCHC3 tissue expression based on datasets of HPA (Human Protein Atlas), GTEx, and FANTOM5 (function annotation of the mammalian genome). (B) Subcellular localization of ZCCHC3. (C) Proteins that interact with ZCCHC3.
Analysis of the relationship between ZCCHC3 expression and prognosis
Using single variate Cox regression analysis, we assessed correlation between the respective expression level of ZCCHC3 and OS in different cancer types, using data from TCGA database. Cox regression identified that high ZCCHC3 expression was a risk factor for LGG, GBMLGG, and LIHC. However, it appeared to be a protective factor in PAAD and KIRC (Figure 4A). Cox regression analysis of DSS identified that high ZCCHC3 expression was a risk factor in LGG and GBMLGG. However, it was a protective factor in KIRC, PAAD, KIPAN, and UCEC (Figure 4B). Cox regression analysis of DFI identified that higher ZCCHC3 expression was a risk factor for PRAD, LUSC, and KIRP (Figure 4C). However, it was a protective factor in ESCA and STES. Cox regression analysis of PFI identified high ZCCHC3 expression as a risk factor in LGG, LIHC, GBMLGG, LUSC, and DLBC, while it was a protective factor in PAAD and KIRC (Figure 4D).

Figure 4: Analysis of the relationship between ZCCHC3 expression and prognosis;(A) Association of ZCCHC3 expression with patient overall survival (OS); (B) Association of ZCCHC3 expression with patient disease‐specific survival (DSS); (C) Association of ZCCHC3 expression with patient disease‐free interval (DFI); (D) Association of ZCCHC3 expression with patient progression-free interval (PFI).
ZCCHC3 and tumor Immune infiltration
We examined the correlation coefficients between ZCCHC3 levels and immune infiltration in various cancer types through the TIMER2.0 database. The results of TIMER 2.0 database showed that the expression of ZCCHC3 was positively correlated with the infiltration levels of Tregs, cancer-associated fibroblast, Dendritic cell, Macrophage, and T cell CD8+ (Figure 5A-E).
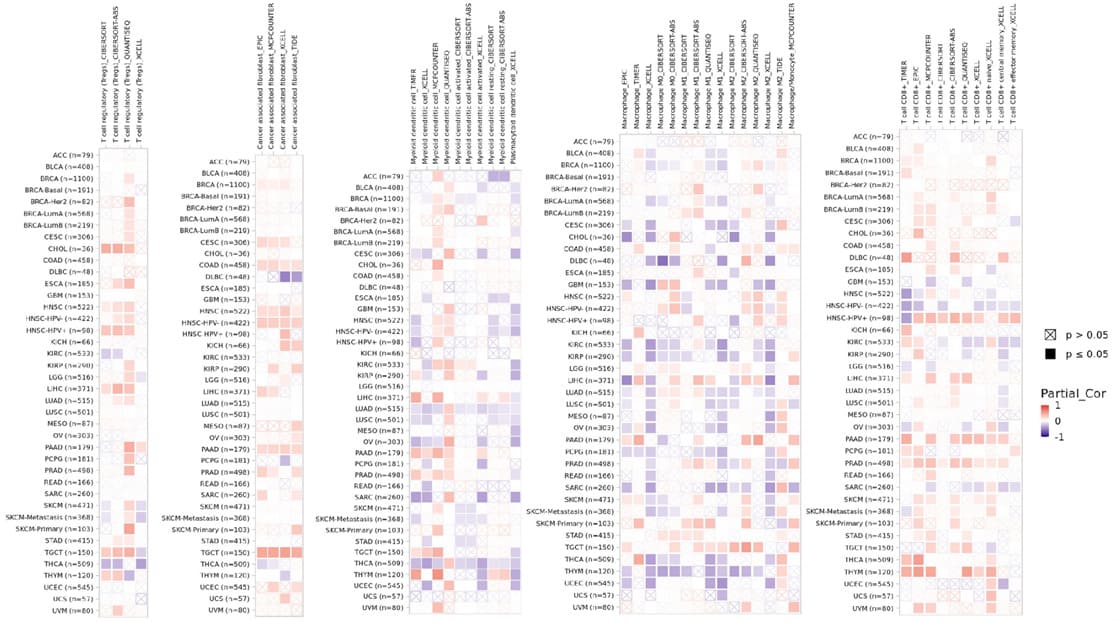
Figure 5: Associations between ZCCHC3 expression and immune infiltrating cells in TIMER2.0 Database; (A) Tregs; (B) Cancer associated fibroblast; (C) Dendritic cell; (D) Macrophage; (E) T cell CD8+.
Relationship between ZCCHC3 and Immune Regulatory Genes
We also investigated the relationship between ZCCHC3 and immune regulatory genes in 33 tumors, including immunostimulator genes, Immunoinhibitor genes, chemokines, chemokines receptors, and MHC (Figure 6 A-E).
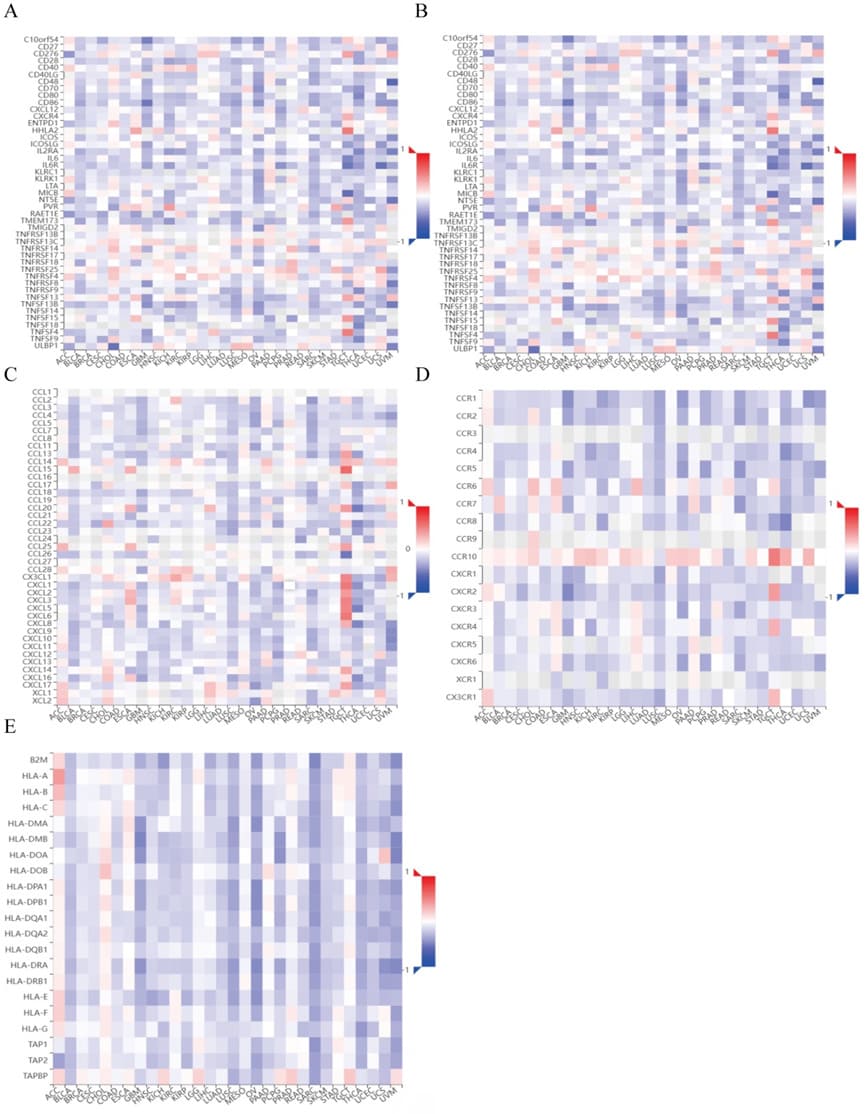
Figure 6: Correlation analysis between ZCCHC3 and immune regulatory genes in pan-cancer; (A) Correlation between ZCCHC3 and immunostimulator genes; (B) Correlation between ZCCHC3 and Immunosuppressor genes; (C) Correlation between ZCCHC3 and chemokines; (D) Correlation between ZCCHC3 and chemokines receptors; (E) Correlation between ZCCHC3 and MHC genes.
In some cancers, ZCCHC3 expression was significantly and positively correlated with immune regulatory genes (Figure 7). Moreover, we found that ZCCHC3 had a positive correlation with immune checkpoints, such as EDNRB, VEGFA, CD276, ADORA2A, IL12A, VEGFB, and ARG1 in a great many cancers (Figure 8).

Figure 7: Correlation analysis between ZCCHC3 and immune regulatory genes in pan-cancer.

Figure 8: Correlation between ZCCHC3 and immune checkpoints.
Analysis of single-cell sequencing results from the TISCH database
Single-cell RNA-Seq data were used to analyze the heterogeneity of ZCCHC3 expression in KIRC, HNSC, LIHC and CHOL (Figure 9 A-D). Unfortunately, The results showed that ZCCHC3 has low cell type specificity.

Figure 9: ZCCHC3 expression analyzed by single-cell sequencing;(A) KIRC; (B) HNSC; (C) LIHC; (D) CHOL.
Correlation between ZCCHC3 Expression and CNV, TMB, MSI in Pan-Cancer
We mapped the mutational profiles of ZCCHC3 across 30 types of cancers (Figure 10A). The results showed that BLCA (1.5%) cancer had the highest mutation rate. We further explored whether ZCCHC3 expression was dependent on the CNV status in the 32 types of cancers, and found statistically significant differences in 17 types of cancers (Figure 10B), including GBMLGG, LGG, CESC, LUAD, COADREAD, BRCA, ESCA, STES, SARC, KIPAN, STAD, PRAD, UCEC, LUSC, LIHC, OV, and UCS. Moreover, the association between TMB / MSI and ZCCHC3 expression was also evaluated. We found that ZCCHC3 expression was positively correlated with TMB in GBMLGG and LGG, but negatively correlated with BRCA, PRAD, THCA, PAAD, UVM, and DLBC (Figure 10C). We also found that the ZCCHC3 level was positively correlated with MSI in CESC, LUAD, ESCA, KIRP, PRAD, KIRC, LUSC, SKCM, BLCA, and KICH, but negatively correlated with GBMLGG and DLBC (Figure 10D).
Discussion
Members of the CCHC-type zinc finger protein family play an important role in the development of cancer. Studies have shown that ZCCHC2 knockdown can promote the proliferation of retinoblastoma cells, while overexpression of ZCCHC2 is on the contrary. ZCCHC2 binds to c-Myc and negatively regulates the polyubiquitination of c-Myc linked to K63, thereby inhibiting the activity of c-Myc and thereby participating in the regulation of the occurrence of retinoblastoma [10]. The expression of ZCCHC3 exacerbates the migration, invasion and epithelialmesenchymal transformation of osteosarcoma cells, which may provide potential guidance for researchers to find effective targets for the treatment of osteosarcoma [8]. ZCCHC4 can promote the growth of liver cancer and participate in the apoptotic resistance of liver cancer to DNA-damaged chemotherapy drugs, and the apoptotic resistance of ZCCHC4 is widespread in lung cancer, pancreatic cancer and colon cancer [11-12]. The expression level of ZCCHC6 in HNSCC patients is significantly increased, and the sensitivity and specificity of diagnosis of HNSCC are 100.00% and 70.83%, respectively [13]. The expression level of ZCCHC6 can be used for HNSCC screening. ZCCHC9 is mainly located in the nucleus, which is consistent with the results previously found that ZCCHC9 is mainly located in the cytoplasm of normal mouse brain and testicular tissue. However, in non-small cell lung cancer (NSCLC) cells, ZCCHC9 is mainly located in the cytoplasm, and its ectopic cytoplasmic expression is significantly correlated with advanced tumor and poor clinical prognosis. Further molecular mechanism studies showed that cytoplasmic ZCCHC9 promoted the proliferation and invasion of NSCLC through the JNK pathway, and treatment with SP600125, an inhibitor of the JNK pathway, showed that the function of ZCCHC9 promoting the proliferation and invasion of cancer cells could be significantly cancelled [1415]. Down-regulation of ZCCHC12 significantly inhibited colony formation, migration and invasion of papillary thyroid cancer cells, suggesting that ZCCHC12 may be a novel molecular marker for PTC diagnosis [16-17]. ZCCHC13 is located in the Xq13.2 region of human X chromosome, which is rich in CT gene, and its expression is upregulated in human hepatocellular carcinoma cells and tissues, which may be related to DNA hypomethylation in promoter region. Overexpression of ZCCHC13 can promote cell cycle progression by promoting G1-S transformation. It is related to the abnormal activation of TK/ERK/c-MYC/CDK pathway, suggesting that ZCCHC13 plays the role of HCC oncogene and can be used as a therapeutic target for HCC [18]. ZCCHC14, with a size of about 100 kDa, is expressed in almost all organs such as human brain, lung and liver. However, recent studies have shown a different situation, showing low or even no expression of ZCCHC14 in human NSCLC tissues, resulting in significantly enhanced cell proliferation and invasion ability.
ZCCHC14 regulates the proliferation and invasion of NSCLC through the P38 pathway. The low expression of ZCCHC14 is significantly correlated with TNM stage, differentiation degree and adverse clinical outcome of tumors, and may become a zinc finger target for clinical treatment [19]. Therefore, we conducted a comprehensive pan-cancer analysis of ZCCHC3 through multiple public databases, mainly in terms of gene expression, mutation and functional mechanisms, and our research contributes to an indepth understanding of the molecular mechanism of ZCCHC3, so as to find new and more effective drug targets for clinical treatment of various tumors.
In this study, we found that the expression level of ZCCHC3 varies in different tumors and is closely related to the prognosis of tumor patients. Therefore, the expression effect of different tumor genes is different, which requires further molecular biology experiments. The occurrence and development of tumors is not only related to the genomic instability and epigenetic changes of tumors themselves, but also related to the tumor microenvironment (TME) as the “fertile soil” for the growth and reproduction of “malignant seeds”, to influence each other and promote the occurrence of tumors [20-21]. The tumor microenvironment is composed of tumor cells, stromal cells (including tumor-associated fibroblasts and mesenchymal stromal cells), immune cells (lymphoid and myeloid cells), and extracellular matrix. Normally, immune cells can recognize and clear abnormal tumor cells, and stromal cells maintain the tissue structure and function of the body [22-23].
However, tumor cells for survival and growth can adopt different strategies, inhibit the immune system of anti-tumor effect, stromal cells and immune cells in the TME through cell interaction or autocrine and paracrine, induce extracellular matrix hardness, blood vessels and lymphatic vessel formation, necrotic area formation and tumor metastasis, abnormal immune cells, fibroblasts produce a large number of immune suppressive factors, such as IL-10, EGF, VEGF, TGF-β to promote tumor growth and metastasis, so immune cells play a vital role in the process of tumor development [24-25]. In this study, ZCCHC3 was positively correlated with the immune infiltration scores of most tumors, and based on TIMER analysis, we found that ZCCHC3 levels were significantly correlated with the degree of infiltration of Tregs, cancer-associated fibroblast, Dendritic cell, Macrophage, and T cell CD8+ . Therefore, high ZCCHC3 expression may influence immune cell interactions, leading to the occurrence of immune escape, promoting tumor progression and affecting patient prognosis. Of course, different types of tumors have different effects on the immune cells of the tumor microenvironment, so we also need to consider other clinical characteristics comprehensively. The above is expected to provide new ideas for cancer immunotherapy, and also further establish the prognostic value of ZCCHC3 in cancer patients.
Tumor immunotherapy is a new treatment method developed in recent years, which has greatly changed the treatment plan of cancer and brought new options to cancer patients [26-27]. At the moment, The most widely studied immune checkpoints are Cytotoxic Tlymphocyte antigen-4 (CTLA-4), Programmed death receptor-1 (PD-1), T cell immunoglobulin and mucin-domain containing-3 (TIM-3), and Lymphocyte activation gene-3 (LAG-3) [28-29]. Immunotherapy based on immune checkpoint inhibitors has led tumor therapy into a new era. Looking for predictive biomarkers is the only way to achieve accurate tumor immunotherapy. Our results show that ZCCHC3 expression is positively correlated with several common immune checkpoints such as EDNRB, VEGFA,CD276, ADORA2A, IL12A, and VEGFB, indicating that ZCCHC3 may be a novel target for tumor immunotherapy. As an emerging biomarker, tumor mutation burden (tumor mutational burden) is increasingly playing its role in predicting the efficacy of tumor immunotherapy. Studies have shown that TMB can be used as a biomarker to improve the efficacy of immunotherapy in non-small cell lung cancer and colorectal cancer. MSI is also an important biomarker in immune checkpoint inhibitors, and highfrequency MSI in colorectal cancer is an independent predictor of clinical features and prognosis [30]. Our study shows that ZCCHC3 is positively correlated with TMB expression in GBMLGG and LGG, and negatively with BRCA, PRAD, THCA, PAAD, UVM, and DLBC. The results of correlation analysis between ZCCHC3 expression and MSI indicated that ZCCHC3 expression in CESC, LUAD, ESCA, KIRP, PRAD, KIRC, LUSC, SKCM, BLCA, and KICH were positively correlated with MSI, and negatively with GBMLGG and DLBC.
We are committed to pan-cancer analysis of ZCCHC3 in the belief that we can find common diagnostic and prognostic markers among tumors and assist in tumor screening, diagnosis, monitoring of tumor recurrence and metastasis, and contribute in the assessment of tumor prognosis and efficacy. The results of this study provide a preliminary basis for understanding the function of ZCCHC3, but it still has some limitations. Through bioinformatics analysis, more experimental data are needed to verify these results and explain the specific biological mechanism of ZCCHC3 in various tumors.
Conclusion
Our research shows that ZCCHC3 has value in the diagnosis and prognosis of a wide range of cancers and regulates cancer through a variety of mechanisms, which may contribute to understanding the role of ZCCHC3 in cancer immunomodulation and immunotherapy.
Acknowledgements
The authors of this manuscript sincerely appreciate the efforts of all researchers who have contributed the data to the public databases of TCGA, GETx, CCLE, TIMER 2.0, HPA database, and sangerbox Online website. The interpretation and reporting of these data are the sole responsibility of the authors.
Statements & Declarations Funding
This work was Supported by the Program of Natural Science Foundation of Zhejiang Province (LY20H160017) , Chinese Medicine Study Foundation of Zhejiang Province (2020ZB292) and PhD research startup foundation of Lishui People’s Hospital (2020bs01).
Conflicts of interest
The authors declare that they have no conflict of interest.
Date Availability
The authors certify that all the original data in this research could be obtained from the public databases.
Author contributions
All authors contributed to the study conception and design.Ming Qin, Junmin Luo and Jihong Feng conceptualized the study; Xuehan Gao, Pinzhi Dong, Linna Wei, Jin Chen, and Haiyan Wang carried out the investigations; Xuehan Gao wrote the original draft; Pinzhi Dong, Linna Wei, Jin Chen, and Haiyan Wang contributed with the writing—reviewing and editing of the paper; Xuehan Gao and Pinzhi Dong visualized the study; Jihong Feng contributed with the funding acquisition.
References
- E Dolgin (2021) Cancer’s new normal. Nat Cancer 2: 1248-1250.
- A Mullard (2020) Addressing cancer’s grand challenges. Nat Rev Drug Discov 19: 825-826.
- N Wang, Zhu L, Wang L, Shen Z, Huang X (2022) Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers. Comput Struct Biotechnol J 20: 3106-3119.
- Wang Y, Yu Y, Pang Y, Yu H, Zhang W, et al. (2021) The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol 18: 2107-2126.
- XN, Chen YD, Fu YZ, Hu MM, Lei CQ, et al. (2018) ZCCHC3 is a cosensor of cGAS for dsDNA recognition in innate immune response. Nat Commun 9: 3349.
- Lian H, Zang R, Wei J, Ye W, Hu MM, et al. (2018) The Zinc-Finger Protein ZCCHC3 Binds RNA and Facilitates Viral RNA Sensing and Activation of the RIG-I-like Receptors. Immunity 49: 438-448.e5.
- Zang R, Lian H, Zhong X, Yang Q, Shu HB (2020) ZCCHC3 modulates TLR3-mediated signaling by promoting recruitment of TRIF to TLR3. J Mol Cell Biol 12: 251-262.
- Wang L (2020) ELF1-activated FOXD3-AS1 promotes the migration, invasion and EMT of osteosarcoma cells via sponging miR-296-5p to upregulate ZCCHC3. J Bone Oncol 26: 100335.
- De Marco C, Zoppoli P, Rinaldo N, Morganella S, Morello M, et al. (2021) Genome-wide analysis of copy number alterations led to the characterisation of PDCD10 as oncogene in ovarian cancer. Transl Oncol 14: 101013.
- Dai H, Yan M, Li Y (2020) The zinc-finger protein ZCCHC2 suppresses retinoblastoma tumorigenesis by inhibiting HectH9-mediated K63linked polyubiquitination and activation of c-Myc. Biochem Biophys Res Commun 521: 533-538.
- Ma H, Wang X, Cai J, Dai Q, Natchiar SK, et al. (2019) N6Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 150: 88-94.
- Zhu H, Chen K, Chen Y, Liu J, Zhang X, et al. (2022) RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct Target Ther 7: 240.
- Arayataweegool A, Srisuttee R, Bin-Alee F, Mahattanasakul P, Tangjaturonrasme N, et al. (2020) Induction of ZCCHC6 expression in peripheral blood mononuclear cells by HNSCC secretions. Gene 754: 144880.
- Zhou A, Zhou J, Yang L, Liu M, Li H, et al. (2008) A nuclear localized protein ZCCHC9 is expressed in cerebral cortex and suppresses the MAPK signal pathway. J Genet Genomics 35: 467-472.
- Shi X, Jiang B, Liu H, Fan C (2019) ZCCHC9 promotes proliferation and invasion of lung cancer through regulating the JNK pathway. J Cell Biochem 120: 10596-10604.
- Li QL, Chen FJ, Lai R, Guo ZM, Luo R, et al. (2012) ZCCHC12, a potential molecular marker of papillary thyroid carcinoma: a preliminary study. Med Oncol 29: 1409-1417.
- Wang O, Zheng Z, Wang Q, Jin Y, Jin W, et al. (2017) ZCCHC12, a novel oncogene in papillary thyroid cancer. J Cancer Res Clin Oncol 143:1679-1686.
- Li Z, Li Z, Wang L, Long C, Zheng Z, et al. (2019) ZCCHC13-mediated induction of human liver cancer is associated with the modulation of DNA methylation and the AKT/ERK signaling pathway. J Transl Med 17: 108.
- Shi X, Han X, Cao Y, Li C, Cao Y (2021) ZCCHC14 regulates proliferation and invasion of non-small cell lung cancer through the MAPK-P38 signalling pathway. J Cell Mol Med 25: 1406-1414.
- L Bejarano, Jordao MJC, Joyce JA (2021) Therapeutic Targeting of the Tumor Microenvironment[J]. Cancer Discov 11: 933-959.
- A Tiwari, Trivedi R, Lin SY (2022) Tumor microenvironment: barrier or opportunity towards effective cancer therapy. J Biomed Sci 29: 83.
- D Chen, Zhang X, Li Z, Zhu B (2021) Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics 11: 1016-1030.
- Elhanani O, Ben-Uri R, Keren L (2023) Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 41: 404-420.
- K Liu, Cui JJ, Zhan Y, Ouyang QY, Lu QS (2022) Reprogramming the tumor microenvironment by genome editing for precision cancer therapy. Mol Cancer 21: 98.
- Di Martino JS, Mondal C, Bravo-Cordero JJ (2019) Textures of the tumour microenvironment. Essays Biochem 63: 619-629.
- Galluzzi L, Humeau J, Buque A, Zitvoge L, Kroemer G, et al. (2020) Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol 17: 725-741.
- J Yang, Zhao S, Wang J, Sheng Q, Liu Q, et al. (2021) A pancancer immunogenomic atlas for immune checkpoint blockade immunotherapy. Cancer Res 82: 539-542.
- Herati RS, Knorr DA, Vella LA, Silva LV, ChilukuriX L, et al. (2022) PD-1 directed immunotherapy alters Tfh and humoral immune responses to seasonal influenza vaccine. Nat Immunol 23: 1183-1192.
- Kennedy LB, Salama AKS (2020) A review of cancer immunotherapy toxicity. CA Cancer J Clin 70: 86-104.
- Y Zhuo, Li S, Hu W, Zhang Y, Shi Y, et al. (2022) Targeting SNORA38B attenuates tumorigenesis and sensitizes immune checkpoint blockade in non-small cell lung cancer by remodeling the tumor microenvironment via regulation of ZCCHC3/AKT/mTOR signaling pathway. J Immunother Cancer 10 :e004113.
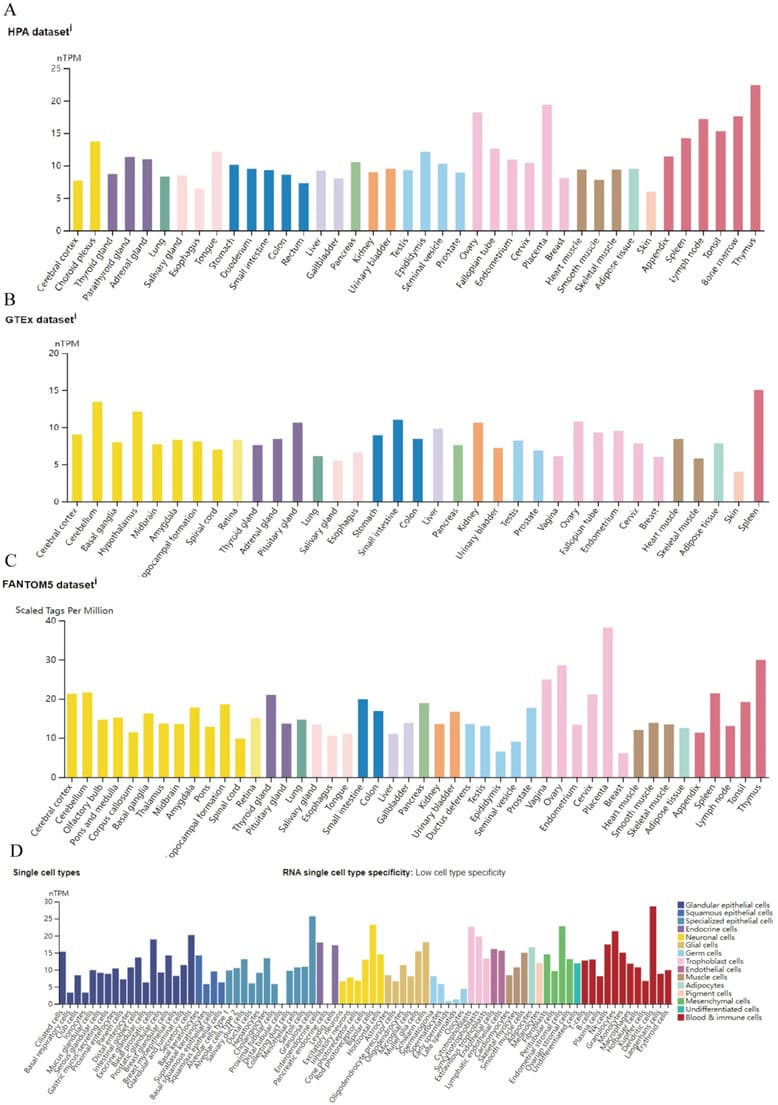
Figure 1: ZCCHC3 expression status in different normal tissues;(A-C) ZCCHC3 tissue expression based on datasets of the HPA (Human protein atlas); GTEx, and FANTOM5 (Function annotation of the mammalian genome 5); (D) ZCCHC3 expression in various cell types.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.