Circrna Expression Profiles and the Role of HDAC9 in Cisplatin Resistance of Esophageal Squamous Cell Carcinoma
by Nanxin Zhu, Jiechun Lin, Lixuan Liu, Zejin Pu, Yu Zhong, Yang Chen, Lingfei Wu*
Department of Gastroenterology, Second Affiliated Hospital, Shantou University Medical College; Shantou, Guangdong515041, P.R. China
*Corresponding author: Lingfei Wu, Department of Gastroenterology, Second Affiliated Hospital, Shantou University Medical College; Shantou, Guangdong515041, P.R. China. Email: 1808435253@qq.com
Received Date: 02 May 2024
Accepted Date: 11 July 2024
Published Date: 13 July 2024
Citation: Nanxin Zhu, Jiechun Lin, Lixuan Liu, Zejin Pu, Yu Zhong, et al. (2024) Circrna Expression Profiles and the Role of HDAC9 in Cisplatin Resistance of Esophageal Squamous Cell Carcinoma. J Surg 9: 11049 https://doi.org/10.29011/2575-9760.11049
Abstract
Background: Cisplatin (CDDP) is one of the standard treatment drugs for esophageal cancer (EC), but recurrence and metastasis are common due to intronic or acquired resistance. Research has shown that circRNAs may be involved in the regulattion of chemotherapy resistance. However, the underlying mechanism is still largely unknown.
Methods: TE-1 (the human Esophageal Squamous Cell Carcinoma, ESCC) cell line that had acquired cisplatin resistance (TE-1/ CDDP) were established by means of the drug concentration gradient increasing method. The half maximal inhibitory concentration of cisplatin (IC50) and the biological characteristics of TE-1 and TE-1/CDDP cells were explored by CCK-8 assay, clone formation and transwell experiments. CircRNA microarray was utilized for identifying the gene expression patterns. Small interfering RNA was prepared to knock down the target gene and its biological effects on drug-resistant were observed. Western blotting was used to assess protein expression.
Results: After 8 months, cisplatin resistant cell line was successfully established. CCK-8 assay indicated that TE-1/CDDP had 8.70fold increased IC50 in comparison with TE-1. Compared with TE-1, TE-1/CDDP cells displayed irregular morphology and exhibited stronger migration and invasion ability than its parental cells. cDNA microarray analysis shown that totally 410 circRNAs were Differentially Expressed (DE). 124 of which were up-regulated while the other 286 were down-regulated in the cisplatin resistant group. Among them, 191 DE host genes were identified, including 56 up-regulated and 135 down-regulated. Functional annotation revealed that the DE circRNAs were mainly involved in p53 signaling pathway, base excision repair, cellular carbon metabolism, Tricarboxylic Acid Cycle (TCA cycle) and propionate metabolism. Histone deacetylase-9 (HDAC9) was three-fold upregulated and was chosen as a cisplatin resistance host gene. Further experimental validation revealed that knockdown of HDAC9 could inhibit proliferation and invasion, as well as the protein expression of PAI-1, AKT, and ERK in TE-1/CDDP cells. HDAC9 appears to confer cisplatin resistance by targeting the AKT and ERK proteins.
Conclusion: Our findings identified multiple aberrantly expressed circRNAs in cisplatin resistance EC cells that may provide a useful resource for identifying novel drug resistance associated circRNAs. The host gene HDAC9 participated in the regulation of cisplatin resistance and might be a therapeutic target in ESCC.
Key words: Cisplatin Resistance, CircRNA, Esophageal Squamous Cell Carcinoma, , Gene Chip, Histone deacetylase-9, TE-1
Introduction
Esophageal Cancer (EC) is still a common disease in the world. According to the latest statistics, approximately 604,000 novel cases and 544,000 mortalities from EC are reported annually worldwide, ranking seventh in incidence, and ranking sixth in overall deaths [1]. East Asia had the highest regional incidence, partly due to the high burden in China, which recorded 278,121 new cases of EC and 257,316 deaths in 2019, accounting for more than half of the global number of cases and deaths [2]. EC has two common histologic types: Esophageal Squamous Cell Carcinoma (ESCC) and Esophageal Adenocarcinoma (EAC) [3]. Most EAC cases occur in developed countries, whereas more than 90% of ESCC cases occur in Asia and sub-Saharan Africa [1]. For advanced esophageal cancer, chemotherapy based on cisplatin (CDDP) is considered as the first-line treatment [4,5]. Although some progress has been achieved in the early diagnosis and treatment, the prognosis for patients with advanced EC remains poor. Cisplatin (CDDP) is commonly used to treat a multitude of tumors including gastric tumors, hepatocellular carcinoma, colon cancer, and esophageal tumors [6-8]. However, patients benefited during the initial 4-6 cycles of chemotherapy and the majority of patients subsequently develop cisplatin resistance [9,10], which is the main reason for chemotherapy failure and the molecular mechanisms are not completely understood [11,12].
Circular RNAs (circRNAs) are a special type of endogenous noncoding RNA that is selectively cleaved and widely expressed in eukaryotic cells. CircRNAs belong to long-chain non-coding RNA (lncRNAs) and play an important role in human diseases [13-16]. Aberrant expression of circRNAs may be associated with tumor development and progression. CircRNAs can be divided into three categories: exonic circular RNA, exon-intron circular RNA, and circular intronic RNA. The main functions of circRNAs are acting as a sponge of microRNAs, binding to RNA binding proteins, affecting the formation of homologous linear mRNAs and regulating transcription. Some circRNAs can also directly code protein, which changed the impression that circRNAs only act as noncoding RNAs [17-19]. Furthermore, studies have shown that circRNA expression profiles differ in different esophageal derived tissues and directly affect tumor cell proliferation and tumor progression [20]. In addition, it has also been reported that circRNAs regulate cisplatin chemotherapy [21]. However, the mechanisms between circRNAs and cisplatin resistance in EC remains unknown. In this study, we established a CDDP resistance model and detected the biological characteristics between the resistant strain (TE-1/CDDP cell) and the parent strain (TE-1 cell). Especifically, we evaluated whether chemotherapy-resistant EC cells show a characteristic circRNA expression pattern, and whether the expression of CDDP resistance-related targets regulated by circRNAs is altered in EC cells. Small interfering RNA technology was used to knock down the target gene (HDAC9) regulated by circRNA and the biological mechanisms related to drug resistance were investigated. Gene chip technology was used to detect the differentially expressed circRNA profiles of the two group cells, identify differentially expressed circRNAs and their regulated target genes.
Materials and Methods
Cell Culture and Establishment of Cell Lines with Acquired Resistance to Cisplatin
TE-1 cells (a human ESCC cell line) were purchased from the Chinese Academy of Sciences in Shanghai, China. All cell lines were validated by STR profiling and tested negative for mycoplasma. Cells were cultured in a humidified incubator at 37 °C and 5% CO2. The culture medium for TE-1 cells consisted of RPMI 1640 supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin (both from Gibco, Thermo Fisher Scientific, Inc., USA). The cisplatin resistant cell model was established by concentration gradient increasing method [22]. In short, the cisplatin resistant cells that grew well in a culture medium containing 0.5 µmol/ml cisplatin were obtained by exposure to cisplatin for 8 months with step-wise escalation of cisplatin concentration. Detection of cisplatin halfmaximal inhibitory concentration (IC50): TE-1 or TE-1/CDDP cells (5,000 cells per well) were seeded into 96-well plates. After treating with different concentrations of cisplatin for 96 hours, the growth rate of these cells was measured using CCK8 methods. Inhibitory rates and the cisplatin resistance index (RI) of TE-1/ CDDP was calculated by Microsoft Excel, and the IC50 values were calculated using GraphPad Prism 8.0 software.
RNA Extraction and Reverse Transcription‑Quantitative PCR (RT‑Qpcr)
To extract the total RNA from cell cultures, Trizol® reagent solution (Invitrogen; Thermo Fisher Scientific, Inc) was used. cDNAs were synthesized using the GoScript Reverse Transcription System RT reagent Kit (Promega Corporation, USA). Next, qPCR was conducted using SYBR Green PCR Master Mix (Promega Corporation) on a Roche Applied Science LightCycler 480II system (Hoffman-La Roche Ltd., Basel, Switzerland). Primers for HDAC9 were designed through Primer 5.0 and synthesized by Realgene (Nanjing, China). The primer sequences for HDAC9 and GAPDH were 5′-AGTAGAGAGGCATCGCAGAGA-3′ and 5′-GGAGTGTCTTTCGTTGCTGAT-3′, 5′-GTCTCCTCTGACTTCAACAGCG-3′ and 5′-ACCACCCTGTTGCTGTAGCC AA-3′, respectively. GAPDH was used as an internal control for normalization. The PCR cycle conditions were an initial denaturation at 95℃ for 10 min. Afterwards the following thermocycling protocol was utilized for 40 cycles: 95°C for 15 sec, 60℃ for 60 sec, and a final extension at 60℃ for 1 min. GAPDH was used as the endogenous controls. The relative expression level of mRNA was calculated by using the 2-ΔΔCq method [23].
Cell Counting Kit‑8 (CCK‑8) Assay
TE-1 cells (5x103 cells/well) were plated into 96-well plates (Corning, Inc.). After incubation at different time intervals, 10 μl CCK-8 reagent (Beyotime Institute of Biotechnology) was added to every well and incubated at 37℃ for 2h. At the indicated time points, the optical density (OD) value was quantified at 450 nm with a microplate reader (Thermo Fisher Scientific, Inc.).
Colony Formation Assay
TE-1 cells (2x103 cells) were seeded into 6-well plates and cultured at 37℃ for 14 days. After visible colonies were formed, the cells were fixed with 4% formaldehyde (Wuhan Servicebio Technology Co., Ltd.) at 37℃ for 20 min. Cell colonies were subsequently stained with 0.1% crystal violet (Sangon Biotech, Co., Ltd.) solution for 20 min and imaged using a light microscope (magnification, x40; Olympus CX31; Olympus Corporation, Tokyo, Japan).
Wound Healing Assay
To conduct the experiment, TE-1 cells were plated onto 6-well plates and allowed to incubate for 24 hours at a density of 2×106 cells per well. When the cells reached full confluence (90-100% coverage), a sterile pipette tip was employed to generate scratches in the cell monolayer. Subsequently, the cells were cultured in a medium devoid of fetal bovine serum (FBS) at a temperature of 37℃. After various time intervals, the cells that migrated into the scratch area were observed using a light microscope (magnification, x100; Olympus CX31; Olympus Corporation). Images were captured and subjected to analysis using ImageJ software (version 1.46; National Institutes of Health, Bethesda, MD, USA).
Transwell Assay
For the migration experiment, TE-1 cells at a concentration of 5x104 cells/well were resuspended in 100 μL RPMI 1640. These cells were subsequently seeded onto an 8-µm pore size chamber (Corning Inc.; Sigma-Aldrich Co.) without any prior coating of Matrigel. The bottom of the chamber was then supplemented with 600 μL of complete medium. Following a 24hour incubation period, the cells that failed to migrate through the membrane of chamber were eliminated. Conversely, the cells that successfully migrated beneath the chamber were fixed with 4% paraformaldehyde (Wuhan Servicebio Technology Co., Ltd.). After fixation, these cells were stained with 0.1% crystal violet (Sangon Biotech, Co., Ltd.) for a duration of 30 minutes at room temperature. Stained cells were counted in three fields using a microscope (magnification, x200; Olympus CorporationCX31; Olympus Corporation, Tokyo, Japan). For invasion assay, the same experimental procedure was followed, with the exception that the chambers were pre-coated with Growth Factor Reduced Matrigel (1:8, Corning, New York, USA).
Microarray and Bioinformatics Analysis
TE-1 cells and TE-1/CDDP cells were hybridized in a hybridization oven for 17 hours at 65℃. The chips were then scanned with a Sure Scan Dx scanner. To normalize the microarray data, we utilized the Linear Models for Micro Array Data package available in the R software. The analysis was conducted on the Affymetrix PrimeViewTM Human Gene Expression Array. The raw data underwent several processing steps including background correction using Robust Multichip Average (RMA), quantile normalization, and probe summarization. This processing was carried out using Expression Console software (version 1.3.1, Affymetrix). To identify differential circRNA expression, we employed ANOVA (analysis of variance) with the p-value adjusted using GeneSpring software (version 14.8, Agilent Technologies). Additionally, we performed Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using the “clusterProfler” and “pathview” software packages[24,25]. The GO analysis can be accessed at http:// www.geneontology.org, while KEGG information can be found at https://www.kegg.jp/. GO includes three categories: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The top 5 enrichment pathways of GO and the first 10 enrichment pathways of KEGG were selected respectively. The circRNA microarray data in this study have been made publicly available in the NCBI Gene Expression Omnibus (GEO) data base under the accession number GSE242834 (http://www.ncbi.nlm. nih.gov/geo/ query/acc.cgi?acc = GSE242834).
Identification of Differentially Expressed Circrnas
The cells were divided into two groups, with each group containing four TE-1 samples and four TE-1/CDDP samples. In datasets, the probes corresponding to multiple circRNAs were removed. Those circRNAs corresponding to multiple probes were averaged as the expression level. The raw data from the two groups were normalized and analyzed. After scale standardization and logarithmic processing of datasets, the “Limma” software package in R software was used to identify differentially expressed circRNAs under the screening criteria of |log2 Fold Change (FC)| ≥ 2 or ≤ 0.5 and p value < 0.05 (a FC ≥ 2 indicated that the gene was 2 times up-regulated, and conversely ≤ 0.5 indicated a half- down-regulated gene). Volcano map and heat map were generated using the “ggplot2” and “pheatmap” packages in R software.
Sirna and Transfection
For RNAi experiments, based on the mRNA sequence of HDAC9 from GenBank and criteria for designing small interfering RNA (siRNA), three si-RNA targeting HDAC9 (si-HDAC9_001, 002, 003) and scrambled control si-RNA (si-NC) were purchased from RiboBio Co., Ltd. (Guangzhou, China). The sequences of si-HDAC9 were as follows: si-HDAC9_001, AGCCACCCTCATGTTACTT; si-HDAC9_002, CATTAGAGGTACCCACAAA; si-HDAC9_003, GGTTTCACAGCAACGCATT. The interference effects were detected using RT-qPCR, and the siRNA with the strongest interference effect was used for subsequent experiments. In transfection experiments, the siRNA (100nM final concentration) were transfected into TE-1/CDDP cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) . The transfected cisplatin-resistance cells (TE-1/CDDP) were divided into 2 groups: si-HDAC9 group (si-HDAC9) and control group (si-NC). 48 hours after transfection, cell pellets were collected and subjected to cell biology experiments and western blotting analysis.
Western Blotting
Proteins were extracted from TE-1/CDDP cells as previously described [26]. Cells transfected with si-HDAC9 or si-NC were collected after 48 h and washed thrice with ice-cold PBS. For the cellular protein lysates, RIPA buffer containing PMSF was used. The lysates were then centrifuged at 12,000 rpm at 4℃ for 10 min. Protein concentration was measured by using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, Ma, USA) following the manufacturer’s instructions. For western blotting assay, the supernatant was boiled for 5 min and 50 μg of protein was electrophoresed on a 10% SDS-PAGE gel and then transferred to PVDF membranes (Millipore, Sigma; USA). This was followed by blocking with 5% nonfat dry milk in Tris-buffered saline with 0.5% Tween-20 for 60 min at room temperature. The membranes were then incubated with primary antibodies. The antibodies used were as follows: HADC9 (1:1000, cat. no. ab109446, Rabbit monoclonal, Abcam), PAI-1 1:1000, cat. no. ab66705, Rabbit monoclonal, Abcam), AKT (1:1000, cat. no. ab38449, Rabbit Abcam), ERK1 (1:1000, ab184699, Rabbit monoclonal, Abcam) and β-actin (1:1000, cat. no. ab8227, Rabbit polyclonal, as loading control) at 4℃ overnight. Blots were detected with Horseradish Peroxidase (HRP)-conjugated goat anti-rabbit or rabbit anti-mouse secondary antibody (ABclonal, Wuhan, China) for 2 h at room temperature. Bands were visualized using an ECL detection system (Thermo Scientific, Waltham, MA, USA). Protein expression was analyzed by the Quantity One software (Bio-Rad, Hercules, CA, USA) and normalized to that of β-actin).
Statistical Analysis
GraphPad Prism 8.0 (GraphPad Software; Dotmatics) and SPSS 21.0 software (IBM Corp.) were used for statistical analyses. All experiments were independently repeated ≥3 times, and the results are presented as the mean ± SD. To compare differences between two groups, a two-sided paired Student’s t-test was employed; while for comparing differences among multiple groups, oneway ANOVA test was conducted followed by Tukey’s post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of a Cisplatin‑Resistant Cell Line (TE‑1/CDDP) and Comparison with their Cellular Biological Characteristics between TE‑1 and TE‑1/CDDP
To explore the mechanisms of CDDP-resistance in EC, treatment of TE-1 cells with conditioned CDDP increased viability of the TE-1 cells after 8 months. The IC50 value of CDDP in TE-1 cells was 4.33 µmol/L and the IC50 in TE-1/CDDP cells was 37.67 µmol/L. The RI of TE-1/CDDP was 8.70 (Figure1A, P < 0.05), indicating the successful cultivation and induction of drug-resistant strains. Under a phase contrast microscope, TE-1 cells display round and regular in shape and uniform in size. However, after induction with cisplatin, TE-1/CDDP cells become narrow and elongated irregular shape, with an increase in black particles and protrusion formation in the cytoplasm (Figure.1B). Furthermore, the colony formation assay indicated that TE-1/CDDP cells had less proliferation capacity (Figure1C). Moreover, Transwell and wound healing assay revealed that TE-1/CDDP cells had the enhanced migration and invasion ability, compared to TE-1 cells (Figure. 1D, E and F).
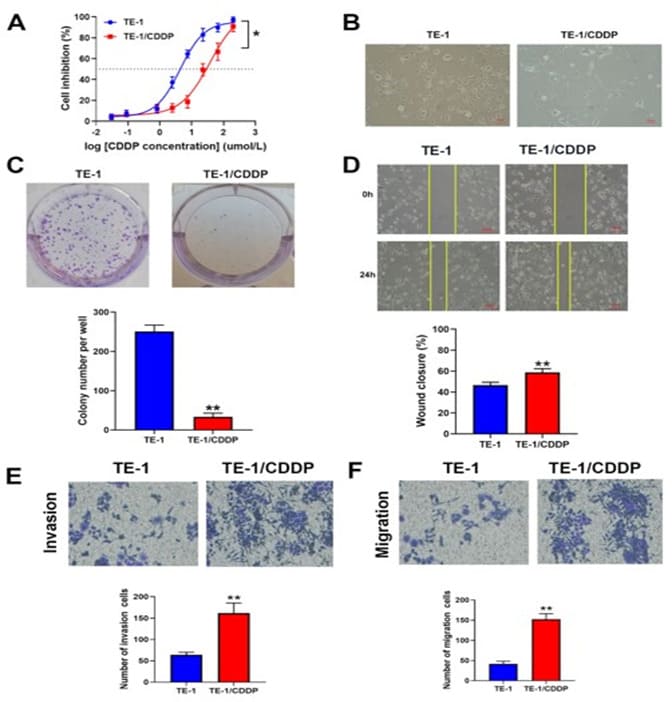
Figure 1: Comparison of cellular biological characteristics between TE-1 and TE-1/CDDP cells. A. IC50 of CDDP was determined by CCK8 assay after treatment with different concentrations of cisplatin on TE-1 or TE-1/CDDP cells. B. The morphology of two group cells was observed under an inverted microscope (magnification, x200; scale bar, 50 µm). C. Cell proliferation was investigated by colony formation assays. D. The graphs display the results of a wound healing assay (magnification, ×200; scale bar, 50 µm). The measurement of wound width was performed using Image J software. E and F. Migration and invasion were assayed using the transwell method between the two group cells. (magnification, ×200) ,*P<0.05 and **P<0.01 in two-tailed Student’s t-test.
Screening for Differentially Expressed Circrnas between TE‑1 and TE‑1/CDDP Cells.
To investigate the effect of circRNAs on EC cells, the circRNAs expression profiles were performed based on the Affymetrix PrimeViewTM Human Gene Expression Array. Scatter plot and volcano plot were used to assess circRNAs expression variation between TE-1 and TE-1/CDDP groups (Figure. 2A-B). These microarray results showed that there were 410 differentially-expressed circRNAs, of which 124 up-regulated and 286 down-regulated. The top 12 up‐regulated and 12 down‐regulated differentially-expressed circRNAs are displayed in heat maps, in which circRNAs are sorted based on differential expression of key host genes (Figure.2C). These differentially-expressed circRNAs were related to 191 host genes (HGs), including 56 up-regulated and 135 down-regulated genes. The top 20 up‐regulated and down‐regulated HGs are displayed in Table 1. Some cisplatin resistance genes, such as LCN2, HDAC9, AREG and EPHA2 showed differential expression in our experiments (Tab.1). HDAC9 was three-fold upregulated, which was related to hsa_circ_0133911, hsa_circ_0005331, hsa_circ_0079527 and hsa_circ_0133904. All the four circRNAs showed significant differential expression and were uploaded to GEO data base (GSE242834). Among these circRNAs, hsa_circ_0133911 had a 3-fold upregulated and was shown in Figure 2C. Thereby HDAC9 was identified as a host gene of circRNAs and chosen for further experiments.
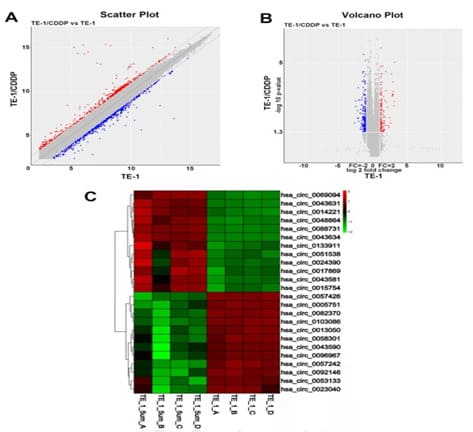
Figure 2. The circRNAs profiling in esophageal cancer cisplatin resistance cells. A. Scatter plots were generated to assess variations in circRNA expression: abscissa represents the signal value normalized to the corresponding data in TE-1. The ordinate represents the normalized signal value of the corresponding data in TE-1/CDDP. B. Volcano plot of circRNAs were constructed by fold change and P-values. Red dots represent genes with increased differential expression, green dots represent genes with decreased differential expression, and gray dots represent insignificant differences in two group cells. C. A heat map was drawn to show the differentially expressed circRNAs. Changes in gene expression levels are shown in a variety of colors, where red represents an increase, blue represents a decrease, and yellow indicates no change. The corresponding degree of change is shown by the depth of color.TE-1-A to TE-1-D are parent strains (TE-1); TE-1-5um-A to TE-1-5um-D are cisplatin resistant strains(TE-1/CDDP(.
|
GeneSymbol |
Fold change |
Up-/down |
GeneSymbol |
Fold change |
Up-/down |
|
regulation |
regulation |
||||
|
LCN2 |
9.024632 |
↑ |
KRT15 |
0.146976 |
↓ |
|
KRT16 |
8.670957 |
↑ |
ITGA4 |
0.167962 |
↓ |
|
KRT17 |
7.991452 |
↑ |
CPA4 |
0.170365 |
↓ |
|
KRT23 |
5.517943 |
↑ |
MPP1 |
0.203346 |
↓ |
|
CFH |
4.200798 |
↑ |
LPHN2 |
0.216551 |
↓ |
|
EML2 |
4.003349 |
↑ |
COL5A2 |
0.226409 |
↓ |
|
C3 |
3.663072 |
↑ |
ARID5B |
0.258988 |
↓ |
|
S100P |
3.606606 |
↑ |
KHK |
0.2644 |
↓ |
|
S100A9 |
3.384165 |
↑ |
GAS2L3 |
0.280337 |
↓ |
|
TAGLN |
3.186655 |
↑ |
STK36 |
0.30541 |
↓ |
|
VIM |
3.127726 |
↑ |
PCSK6 |
0.307323 |
↓ |
|
HDAC9 |
3.006195 |
↑ |
PC |
0.311212 |
↓ |
|
MAP1B |
2.959089 |
↑ |
POLE |
0.315011 |
↓ |
|
EPHA2 |
2.88712 |
↑ |
RNFT2 |
0.325627 |
↓ |
|
AREG |
2.683702 |
↑ |
FASN |
0.333755 |
↓ |
|
FAM129A |
2.477368 |
↑ |
DIS3L2 |
0.335346 |
↓ |
|
ZNFX1-AS1 |
2.44769 |
↑ |
SEMA3C |
0.337317 |
↓ |
|
LAMB3 |
2.436187 |
↑ |
KANK4 |
0.338178 |
↓ |
|
ICAM1 |
2.432076 |
↑ |
DGCR8 |
0.34631 |
↓ |
|
NEBL |
2.423578 |
↑ |
STT3A |
0.353904 |
↓ |
|
Fold-change > 2.0 or < 0.5; P < 0.05. histone deacetylase 9, HDAC9 |
|||||
Summary of circRNA microarray results for the cisplatin resistant cell line (TE-1/CDDP), including dysregulated host genes, fold changes, and regulation in expression, compared to parental cell line (TE-1). ↑: Upregulation of the circRNA; ↓: Downregulation of the circRNA .
Table 1: Top 20 up-regulated and down-regulated host genes
Gene Ontology (GO) enrichment analysis
Gene product annotations were divided into three categories: Biological Process (BP), Molecular Functions (MF), and Cellular Component (CC). GO enrichment analysis revealed that in cisplatin resistant cells, the most significantly BP were negative regulation of B cell activation, spindle localization, and intermediate filament cytoskeleton organization. Differential genes involved in CC were mainly manifested in mitotic spindle, stress fiber, spindle microtubule. Differential genes involved in MF were mainly in scaffold protein binding, microtubule binding, and structural constituent of cytoskeleton. According to the secondary classification annotation map, the host genes involved in biological processes are mainly distributed in the cell growth process (80.62%), single-cell biological growth process (69.63%), biological regulation (57.59%), metabolic process (56.54%) and biological process regulation (55.50%) (Figure. 3A and 3B). Table 2 showed cisplatin-resistance related pathways in the GO analysis, mainly including negative regulation of B cell activation, intermediate filament-based process, mitotic spindle, spindle microtubule, microtubule binding, microtubule motor activity. Through GO enrichment analysis, we found that DEGs play a role in cell growth process, biological regulation, and metabolic process, and differential expression of genes may cause disorders of the above biological functions.
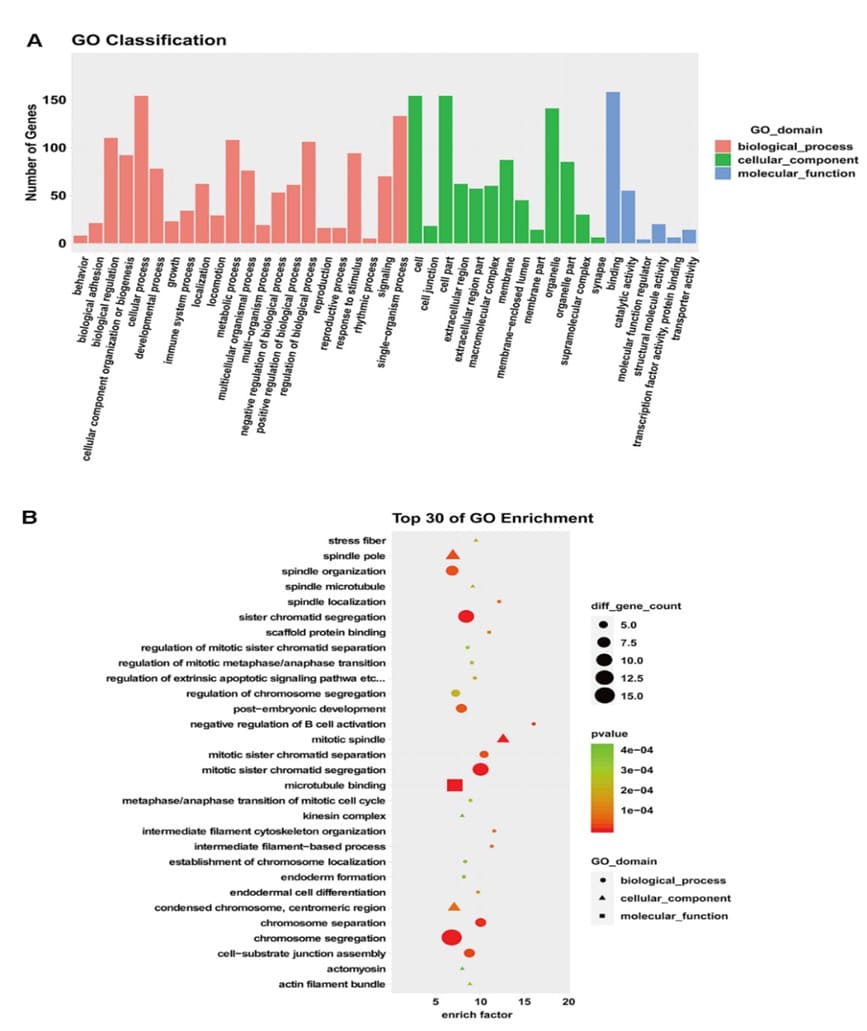
Figure 3: GO enrichment analysis.A. GO classification annotation map: X-axis is the GO classification and Y-axis is the number of genes. B. GO analysis bubble diagram: the ordinate on the left side is the GO function, and the abscissa represents enrichment degree.
|
GO ID |
Term |
Ontology |
Enrichment factor |
P-value |
|
GO:0050869 |
negative regulation of B cell activation |
BP |
15.99 |
0.00002 |
|
GO:0045104 |
intermediate filament cytoskeleton organization |
BP |
11.53 |
0.000084 |
|
GO:0045103 |
intermediate filament-based process |
BP |
11.27 |
0.000093 |
|
GO:0051306 |
mitotic sister chromatid separation |
BP |
10.33 |
0.000031 |
|
GO:0051304 |
chromosome separation |
BP |
9.92 |
0.000009 |
|
GO:0072686 |
mitotic spindle |
CC |
12.61 |
0.000002 |
|
GO:0005876 |
spindle microtubule |
CC |
9.18 |
0.000229 |
|
GO:0005871 |
kinesin complex |
CC |
8 |
0.000419 |
|
GO:0000779 |
condensed chromosome, centromeric region |
CC |
7.08 |
0.000073 |
|
GO:0000922 |
spindle pole |
CC |
6.94 |
0.000027 |
|
GO:0008017 |
microtubule binding |
MF |
7.15 |
0.000007 |
|
GO:0003777 |
microtubule motor activity |
MF |
5.83 |
0.000001 |
|
GO:0015631 |
tubulin binding |
MF |
5.73 |
0.000007 |
|
GO:0001948 |
glycoprotein binding |
MF |
4.72 |
0.004009 |
|
GO:0047485 |
protein N-terminus binding |
MF |
4.51 |
0.004865 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Table 2: GO (BP, CC and MF) enrichment analysis
KEGG Pathway Enrichment Analysis
In KEGG pathway enrichment analysis, carbon metabolism, p53 signaling pathway, base excision repair, TCA cycle, pyrimidine metabolism, and DNA replication were markedly enriched (Table 3). These potential molecular mechanisms provide the basis for new treatment methods of drug resistance. The enriched analysis data were then converted into visual representations using an R program containing the ggplot2 package (Figure. 4A and 4B). KEGG analysis showed that these differential host genes on target pathways were significantly correlated with DNA replication, base exception repair, p53 signaling pathway, cell cycle and pyrimidine metabolism. Among them, ATM, SERPINE1, XRCC1 and UPP1 have been reported as genes involved in drug resistance and cancer metastasis.
|
Pathway name |
P-value |
Count |
EnrichFactor |
Genes |
|
Carbon metabolism |
8.99E-05 |
7 |
5.92 |
PC, SUCLG2, PCCA, ALDOC, G6PD, HIBCH, SDHA |
|
p53 signaling pathway |
0.000296 |
5 |
6.74 |
ATM, SERPINE1, CCNB2, GTSE1, RRM2 |
|
Citrate cycle (TCA cycle) |
0.000669 |
3 |
9.65 |
PC, SUCLG2, SDHA |
|
Propanoate metabolism |
0.000845 |
3 |
9.07 |
SUCLG2, PCCA, HIBCH |
|
Base excision repair |
0.000845 |
3 |
9.07 |
POLE, LIG1, XRCC1 |
|
Pentose phosphate pathway |
0.008404 |
2 |
6.43 |
ALDOC, G6PD |
|
Fructose and mannose metabolism |
0.01083 |
2 |
5.87 |
KHK, ALDOC |
|
DNA replication |
0.01364 |
2 |
5.39 |
POLE, LIG1 |
|
Pyrimidine metabolism |
0.006163 |
3 |
5.25 |
UPP1, TK1, RRM2 |
Table 3: KEGG pathway enrichment analysis
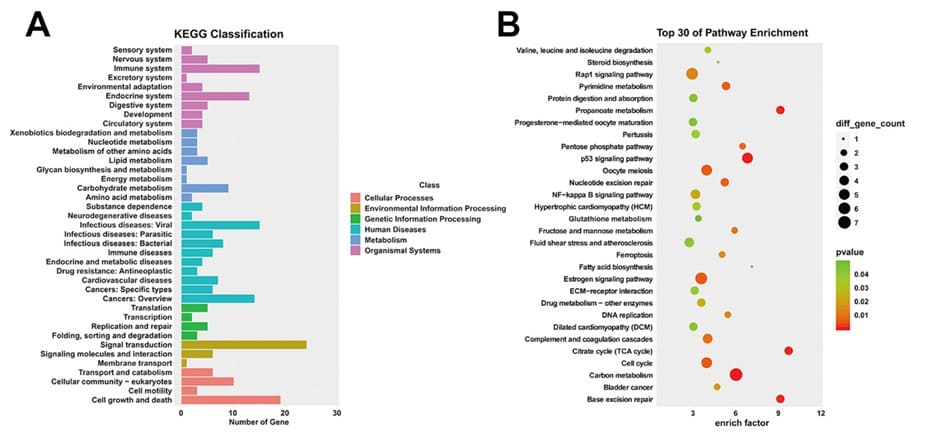
Figure 4. KEGG pathway enrichment analysis. A. KEGG classification annotation map of differential host genes regulated by circRNAs: ordinate indicates the specific signaling pathway and abscissa indicates the number of genes enriched within that pathway. B. Top 30 of KEGG pathway enrichment analysis: a dot in the figure represents a pathway, the X-axis indicates the magnitude of the enrichment, and the left side of the Y-axis marks the specific signaling pathway. The higher the enrichment degree of differential host genes on target pathways, the more reliable the enrichment significance.
Knockdown of HDAC9 Suppresses Cell Proliferation, Invasion and the Protein Expression of PAI‑1, AKT and ERK on TE‑1/ CDDP Cells
HDAC9 has a high differential fold in our differentially expressed gene profile and also involved in a variety of drug resistance related biological functions, we therefore selected HDAC9 for further studies. To further understand the role of HDAC9 in EC cell biological function, three siRNA targeting HDAC9 (siHDAC9_001, 002, 003) were transiently transfected into TE-1/ CDDP cells. RT-qPCR results showed that the relative mRNA expression of HDAC9 was 62%, 45% and 27%, respectively. Of which, si-HDAC9_003 showed the highest interference efficiency and was chosen as the next interference experiments. (Figure. 5A, p< 0.01). To determine whether HDAC9 could be a potential target for esophageal cancer cisplatin resistance, the tumor cell growth, migration and invasion was detected after silencing endogenous HDAC9 expression. As shown in Figure 5C, knockdown of HDAC9 markedly inhibited TE-1/CDDP cell viability. Colony formation assay also confirmed that knockdown of HDAC9 could decrease the colony formation ability on TE-1/CDDP cells (Figure. 5E, p< 0.01). Transwell and wound healing assays validated that knockdown of HDAC9 suppressed TE-1/CDDP cell invasion and migration (Fig. 5B, p< 0.05 and Fig. 5D, p< 0.01). Collectively, these results suggested that knockdown of HDAC9 inhibited cell proliferation and motility in cisplatin resistant TE-1 cells.
In this study, HDAC9 were significantly upregulated, and KEGG enrichment analyses indicated that p53 signaling pathway included ATM, SERPINE1, CCNB2, GTSE1, and RRM2 (Table.3). It is reported that PAI-1, encoded by the SERPINE1 gene, is highly expressed in various tumor tissues, and participates in cancer development [27]. To explore whether HDAC9 increased cell proliferation and invasion was related to PAI-1, the expression levels of PAI-1, AKT and ERK were examined by western blotting. Our results demonstrated that the relative expressions of PAI-1, AKT and ERK were all lower in si-HDAC9 groups in comparison with the si-NC group (p< 0.01, Figure.5F). These results showed that knockdown of HDAC9 inhibits the expression of PAI-1, AKT, and ERK in cisplatin resistant cells.
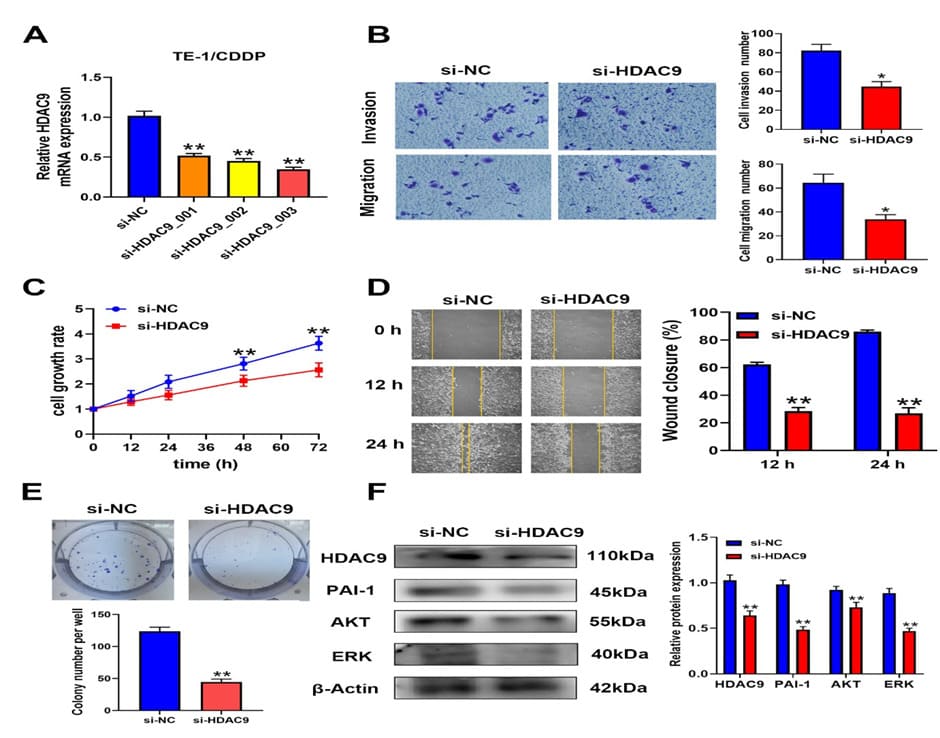
Figure 5. Knockdown of HDAC9 inhibited cell proliferation, invasion and related protein expression in TE/CDDP cells. A. TE-1/CDDP cells were transfected with si-RNA targeting HDAC9 (si-HDAC9_001, 002, 003 and control si-NC). HDAC9 mRNA expression levels were assessed by RT-qPCR. B. Transwell assay was performed to measure the invasion and migration ability of TE-1/CDDP cells. C. CCK8 assay was used to detect the proliferation of TE-1/CDDP cells at different time points D. Wound closure was measured at different time points by scratch wound-healing assay. E. Colony formation of si-NC cells and si-HDAC9 cells. F. Relative protein expression levels of PAI-1, AKT, and ERK were measured by western blotting. Protein’s relative quantity was calculated by normalizing to β-actin. *P<0.05 and **P<0.01 compared with si-NC group in the two-tailed Student’s t-test.
Discussion
Esophageal squamous cell carcinoma is a common malignant tumor in China. At present, more and more ESCC patients have chosen chemotherapy plus immunotherapy for neoadjuvant treatment in order to improve the prognosis. However, the resistant of chemotherapeutic drugs greatly reduces the efficacy. Current research has identified that multiple mechanisms were involved in cisplatin resistance, such as inhibiting cell apoptosis, altering cell cycle checkpoints, and increasing DNA damage repair [28]. In recent years, the effect of circRNAs on tumor resistance to chemotherapy has drawn attention [29,30]. There is increasing evidence that circRNAs are involved in the development of various tumors and closely associated with chemoresistance [31]. Fan et al. demonstrated that circ_0031242 upregulates POU3F2 expression via competitive binding to miR-924, leading to the induction of cisplatin resistance in hepatocellular carcinoma cells [32]. Luo et al also reported that circRNA_101505 could sensitize HCC cells to cisplatin by sponging miR-103, and thereby promoting oxidored-nitro domain-containing protein 1 (NOR1) expression [21]. In addition, circVAPA upregulates STAT3 protein expression by binding to miR-125b-5p, causing cisplatin resistance in gastric cancer [33]. In the current study, first, we successfully established a cisplatin resistant model of esophageal cancer. We found that at the IC50 level, the resistance of TE-1/CDDP against cisplatin was 8.70-fold stronger than that of parental TE-1. Cisplatin resistant cells, although slowing down in growth, had stronger migration and invasion abilities, demonstrating stronger survival ability in the presence of cisplatin. Next, circRNA microarray technology was utilized to evaluate global circRNA expression profiling and 410 differentially expressed circRNAs were screened in the two sorts of cells.
Among 191 host genes, multiple drug resistance genes have been identified, including LCN2, which controls the activation of the NF-κB pathway in oral squamous cell carcinoma [34]; HDAC9, which obstructs the cell apoptosis in gastric cancer[35]; AREG, which affects the AREG-EGFR-ERK pathway in human chondrosarcoma [36]; and EPHA2, which regulates the RSKEphA2-GPRC5A signaling pathway in ovarian cancer [37]. Miller et al. reported that microtubule proteins regulates drug resistance through mitosis, cell migration, and intracellular trafficking [38]; Henriques et al. demonstrated that mitosis participates in the regulation of tumor progression and drug resistance [39]. In our study, GO enrichment analysis showed there were significant differences in both microtubule proteins and mitosis in two group cells, which was consistent with the above-mentioned results.KEGG enrichment analysis showed 191 host genes mainly enriched in p53 signaling pathway, Base Exception Repair (BER), TCA cycle, and pyrimidine metabolism signal pathway. The p53 signaling pathway regulated multiple factors responsible for the drug resistance, such as Nrf2 and AZD-7762 [40]. CDDP exhibits direct cytotoxicity to tumor cells through binding to the nuclear DNA [41,42]. Both eukaryotic and prokaryotic cells are hypersensitive to CDDP because of deficiencies in DNA repair [43]. The enhancement of DNA repair in tumor cells led to resistance to CDDP and subsequent tumor recurrence [44]. The sensitivity and resistance of cancer cells to cisplatin chemotherapy depend on DNA damage response activity. Some studies have shown that BER has a protective effect against platinum-induced cytotoxicity [45]. Jiang et al. reported that targeting UBE2T improves the efficacy of gemcitabine in the treatment of pancreatic cancer by regulating pyrimidine metabolism and replication stress [46]. These signal pathways have also been identified in our experiment. In the study, four proteins, UPP1, XRCC1, ATM and SERPINE1 were identified as p53 signaling pathway in KEGG enrichment analysis. Guan et al. reported that UPP1 overexpression is associated with lymph node metastasis, tumor stage in thyroid cancer. And silencing of UPP1 could inhibit the migration, invasion and proliferation of thyroid cancer cells [47]. Low expression of XRCC1 is associated with poorer disease-free-survival in gastric cancer patients [48]. Furthermore, Li et al. demonstrated that XRCC1 expression could inhibit the invasion, and promoted cell apoptosis, thus inhibiting breast cancer progression [49]. Bueno et al. reported that the downregulation of ATM is associated with poor prognosis in breast cancer [50]. Rui et al. found that SERPINE1 is closely associated with poor prognosis in ESCC patients [51]. The overexpression of SERPINE1 appeared to be associated with N-staging and might be related to angiogenesis and metastasis. Knockdown of SERPINE1 in oral cancer cells resulted in diminished cell proliferation and invasion and increased sensitivity to bleomycin and docetaxel [52]. The above results indicate that the p53 signaling pathway plays an important role in tumor drug resistance, which is consistent with the results of our circRNA gene chip microarray in TE-1/CDDP cells.
PAI-1, encoded by the SERPINE1 gene, is highly expressed in various tumor tissues, and participates in cancer development [27]. Rui et al. found that SERPINE1 is closely associated with poor prognosis in ESCC patients by survival analysis [51]. In addition, PAI-1 is highly expressed in esophageal squamous carcinoma [53], and could promote invasion and migration of ESCC cells via activation of AKT and ERK pathways[54]. In esophageal carcinoma, CDDP could activate cancer-associated fibroblasts to excrete PAI-1, resulting in carcinoma progression and chemoresistance [55]. These evidences suggested that PAI1, AKT, ERK might participate in the regulation of cisplatin resistance in esophageal cancer. In previous studies, HDAC9 was highly expressed in tumor tissues and could act as an oncogene by interacting with transcriptional repressors and oncogenic proteins [56], regulating oncogene acetylation [57], activating JAK/STAT3 signaling pathway [58], promoting angiogenesis [59] and modulating immune cells [60]. HDAC9 also played an important role in various type of cancers, such as lung cancer [61], hepatocellular carcinoma[62], gastric cancer [63],and oral squamous cancer [64].To further elucidate the role of HDAC9 in drug resistance, small interfering RNA technology was utilized. Our results showed that knocking down of HDAC9 inhibited the proliferation in TE-1/CDDP cells. The scratch wound-healing and transwell assays further demonstrated that knockdown of HDAC9 inhibited migration and invasion in cisplatin resistant cells. These results are compatible with previous reports [65,66]. A recent study demonstrated that HDAC9 facilitates cell proliferation and invasion of oxaliplatin-resistant cells in hepatocellular carcinoma [67]. In this study, western blotting assay indicated that knockdown of HDAC9 decreased the protein expression of PAI-1, AKT, and ERK in TE-1/CDDP cells. These findings indicated that HDAC9 could increase cell proliferation, invasion and the effects may be related to PAI-1, AKT and ERK proteins in esophageal cancer. The detailed molecular mechanisms of regulating cisplatin resistance between HDAC9 and related signal pathway need to be further investigated.
Conclusion
In conclusion, we successfully constructed a cisplatin-resistant cell line. Gene chip microarray showed that multiple aberrantly expressed circRNAs participated in regulation of cisplatin resistance in ESCC. HDAC9 was involved in the regulation of cisplatin resistance and might be a therapeutic target for cisplatin resistance in esophageal cancer.
Funding
This study was supported by Natural Science Foundation of Guangdong Province (2021A1515012127).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
NXZ and JCL performed the experimental work; YZ and YC performed the bioinformatic analyses; LXL and ZJP provided reagents and assisted with the cell biology experiments; NXZ, JCL, YZ, YC and LFW analyzed the data and contributed to writing of the manuscript; LFW designed and supervised the study. All authors read and approved the final manuscript.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians 71: 209-249.
- Ma L, Li X, Wang M, Zhang Y, Wu J, et al. (2022) The Incidence, Mortality, and DALYs Trends Associated with Esophageal Cancer - China, 1990-2019. China CDC weekly 4: 956-961.
- Spechler SJ (2013) Barrett esophagus and risk of esophageal cancer: a clinical review. JAMA 310: 627-636.
- Su LL, Chang XJ, Zhou HD, Hou LB, Xue XY (2019) Exosomes in esophageal cancer: A review on tumorigenesis, diagnosis and therapeutic potential. World journal of clinical cases 7: 908-916.
- Florea AM, Büsselberg D (2011) Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers 3: 1351-1371.
- Romani A (2022) Cisplatin in cancer treatment. Biochemical pharmacology 206: 115323.
- Zhang C, Xu C, Gao X, Yao Q (2022) Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 12: 2115-2132.
- Leng X, He W, Yang H, Chen Y, Zhu C, et al. (2021) Prognostic Impact of Postoperative Lymph Node Metastases After Neoadjuvant Chemoradiotherapy for Locally Advanced Squamous Cell Carcinoma of Esophagus: From the Results of NEOCRTEC5010, a Randomized Multicenter Study. Annals of surgery 274: e1022-e1029.
- Shi H, Mao Y, Ju Q, Wu Y, Bai W, et al. (2018) C-terminal binding protein-2 mediates cisplatin chemoresistance in esophageal cancer cells via the inhibition of apoptosis. International journal of oncology 53: 167-176.
- Huang XP, Li X, Situ MY, Huang LY, Wang JY, et al. (2018) Entinostat reverses cisplatin resistance in esophageal squamous cell carcinoma via down-regulation of multidrug resistance gene 1. Cancer letters 414: 294-300.
- Zou FW, Yang SZ, Li WY, Liu CY, Liu XH, et al. (2020) circRNA_001275 upregulates Wnt7a expression by competitively sponging miR-370-3p to promote cisplatin resistance in esophageal cancer. International journal of oncology 57: 151-160.
- Lohan-Codeço M, Barambo-Wagner ML, Nasciutti LE, Ribeiro Pinto LF, Meireles Da Costa N, et al. (2022) Molecular mechanisms associated with chemoresistance in esophageal cancer. Cellular and molecular life sciences : CMLS 79: 116.
- Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA biology 12: 381-388.
- Huang C, Shan G (2015) What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription 6: 61-64.
- Panda AC (2018) Circular RNAs Act as miRNA Sponges. Advances in experimental medicine and biology 1087: 67-79.
- Zheng Y, Li Z, Wang Y, Chen W, Lin Y, et al. (2023) CircRNA: A new class of targets for gastric cancer drug resistance therapy. Pathology oncology research : POR 29: 1611033.
- Kristensen LS, Andersen MS, Stagsted L, Ebbesen KK, Hansen TB, et al. (2019) The biogenesis, biology and characterization of circular RNAs. Nature reviews. Genetics 20: 675-691.
- Li Q, Wang Y, Wu S, Zhou Z, Ding X, et al. (2019) CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell metabolism 30: 157-173.e7.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333-338.
- Ju C, He J, Wang C, Sheng J, Jia J, et al. (2022) Current advances and future perspectives on the functional roles and clinical implications of circular RNAs in esophageal squamous cell carcinoma: more influential than expected. Biomarker research 10: 41.
- Luo YW,Fu YF,Huang R,Gao M, Liu FX, et al.(2019) CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR - 103 and promotes oxidored - nitro domain - containing protein 1 expression.Cell Death Dis 5: 121-130.
- Toshimitsu H, Hashimoto K, Tangoku A, Iizuka N, Yamamoto K, et al. (2004) Molecular signature linked to acquired resistance to cisplatin in esophageal cancer cells. Cancer letters 211: 69-78.
- Li MK, Liu LX, Zhang WY, Zhan HL, Chen RP, et al. (2020) Long non-coding RNA MEG3 suppresses epithelial-to-mesenchymal transition by inhibiting the PSAT1-dependent GSK-3β/Snail signaling pathway in esophageal squamous cell carcinoma. Oncology reports 44: 2130-2142.
- Luo W, Brouwer C (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics (Oxford, England) 29: 1830-1831.
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : a journal of integrative biology 16: 284-287.
- Zhang WY, Zhan HL, Li MK, Wu GD, Liu Z, et al. (2022) Long noncoding RNA Gas5 induces cell apoptosis and inhibits tumor growth via activating the CHOP-dependent endoplasmic reticulum stress pathway in human hepatoblastoma HepG2 cells. Journal of cellular biochemistry 123: 231-247.
- Iwaki T, Urano T, Umemura K (2012) PAI-1, progress in understanding the clinical problem and its aetiology. British journal of haematology 157: 291-298.
- Xu XF, Yang XK, Song Y, Chen BJ, Yu X, et al. (2022) Dysregulation of non-coding RNAs mediates cisplatin resistance in hepatocellular carcinoma and therapeutic strategies. Pharmacological research 176: 105906.
- Xu T, Wang M, Jiang L, Ma L, Wan L, et al. (2020) CircRNAs in anticancer drug resistance: recent advances and future potential. Molecular cancer 19: 127.
- Guo X, Gao C, Yang DH, Li S (2023) Exosomal circular RNAs: A chief culprit in cancer chemotherapy resistance. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy 67: 100937.
- Tao X, Shao Y, Yan J, Yang L, Ye Q, et al. (2021) Biological roles and potential clinical values of circular RNAs in gastrointestinal malignancies. Cancer biology & medicine 18: 437-457.
- Fan W, Chen L, Wu X, Zhang T (2021) Circ_0031242 Silencing Mitigates the Progression and Drug Resistance in DDP-Resistant Hepatoma Cells by the miR-924/POU3F2 Axis. Cancer management and research 13: 743-755.
- Deng P, Sun M, Zhao WY, Hou B, Li K, et al. (2021) Circular RNA circVAPA promotes chemotherapy drug resistance in gastric cancer progression by regulating miR-125b-5p/STAT3 axis. World journal of gastroenterology 27: 487-500.
- Huang Z, Zhang Y, Li H, Zhou Y, Zhang Q, et al. (2019) Vitamin D promotes the cisplatin sensitivity of oral squamous cell carcinoma by inhibiting LCN2-modulated NF-κB pathway activation through RPS3. Cell death & disease 10: 936.
- Xiong K, Zhang H, Du Y, Tian J, Ding S (2019) Identification of HDAC9 as a viable therapeutic target for the treatment of gastric cancer. Experimental & molecular medicine 51: 1-15.
- Huang YW, Lin CY, Tsai HC, Fong YC, Han CK, et al. (2020) Amphiregulin promotes cisplatin chemoresistance by upregulating ABCB1 expression in human chondrosarcoma. Aging 12: 9475-9488.
- Moyano-Galceran L, Pietilä EA, Turunen SP, Corvigno S, Hjerpe E, et al. (2020) Adaptive RSK-EphA2-GPRC5A signaling switch triggers chemotherapy resistance in ovarian cancer. EMBO molecular medicine 12: e11177.
- Kanakkanthara A, Miller JH (2013) MicroRNAs: novel mediators of resistance to microtubule-targeting agents. Cancer treatment reviews 39: 161-170.
- Henriques AC, Ribeiro D, Pedrosa J, Sarmento B, Silva P, et al. (2019) Mitosis inhibitors in anticancer therapy: When blocking the exit becomes a solution. Cancer letters 440-441: 64-81.
- Cao X, Hou J, An Q, Assaraf YG, Wang X (2020) Towards the overcoming of anticancer drug resistance mediated by p53 mutations. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy 49: 100671.
- Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, et al. (2012) Molecular mechanisms of cisplatin resistance. Oncogene 31: 18691883.
- Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, et al. (2014) Systems biology of cisplatin resistance: past, present and future. Cell death & disease 5: e1257.
- Fraval HN, Rawlings CJ, Roberts JJ (1978) Increased sensitivity of UV-repair-deficient human cells to DNA bound platinum products which unlike thymine dimers are not recognized by an endonuclease extracted from Micrococcus luteus. Mutation research 51: 121-132.
- Turner KM, Sun Y, Ji P, Granberg KJ, Bernard B, et al. (2015) Genomically amplified Akt3 activates DNA repair pathway and promotes glioma progression. Proceedings of the National Academy of Sciences of the United States of America 112: 3421-3426.
- Slyskova J, Sabatella M, Ribeiro-Silva C, Stok C, Theil AF, et al. (2018) Base and nucleotide excision repair facilitate resolution of platinum drugs-induced transcription blockage. Nucleic acids research 46: 9537-9549.
- Jiang X, Ma Y, Wang T, Zhou H, Wang K, et al. (2023) Targeting UBE2T Potentiates Gemcitabine Efficacy in Pancreatic Cancer by Regulating Pyrimidine Metabolism and Replication Stress. Gastroenterology 164: 1232-1247.
- Guan Y, Bhandari A, Zhang X, Wang O (2019) Uridine phosphorylase 1 associates to biological and clinical significance in thyroid carcinoma cell lines. Journal of cellular and molecular medicine 23: 7438-7448.
- Wang W, Yang J, Yu Y, Deng J, Zhang H, et al. (2020) Expression of JWA and XRCC1 as prognostic markers for gastric cancer recurrence. International journal of clinical and experimental pathology 13: 31203127.
- Li Q, Ma R, Zhang M (2018) XRCC1 rs1799782 (C194T) polymorphism correlated with tumor metastasis and molecular subtypes in breast cancer. OncoTargets and therapy 11: 8435-8444.
- Bueno RC, Canevari RA, Villacis RA, Domingues MA, Caldeira JR, et al. (2014) ATM down-regulation is associated with poor prognosis in sporadic breast carcinomas. Annals of oncology : official journal of the European Society for Medical Oncology 25: 69-75.
- Zhou R, Liu D, Zhu J, Zhang T (2020) Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis. Medicine 99: e22434.
- Guo X, Sun Z, Chen H, Ling J, Zhao H, et al. (2023) SERPINE1 as an Independent Prognostic Marker and Therapeutic Target for NicotineRelated Oral Carcinoma. Clinical and experimental otorhinolaryngology 16: 75-86.
- Zhang Y, Zhang YL, Chen HM, Pu HW, Ma WJ, et al. (2014) Expression of Bmi-1 and PAI-1 in esophageal squamous cell carcinoma. World journal of gastroenterology 20: 5533-5539.
- Sakamoto H, Koma YI, Higashino N, Kodama T, Tanigawa K, et al. (2021) PAI-1 derived from cancer-associated fibroblasts in esophageal squamous cell carcinoma promotes the invasion of cancer cells and the migration of macrophages. Laboratory investigation; a journal of technical methods and pathology 101: 353-368.
- Che Y, Wang J, Li Y, Lu Z, Huang J, et al. (2018) Cisplatin-activated PAI-1 secretion in the cancer-associated fibroblasts with paracrine effects promoting esophageal squamous cell carcinoma progression and causing chemoresistance. Cell death & disease 9: 759.
- Yang C, Croteau S, Hardy P (2021) Histone deacetylase (HDAC) 9: versatile biological functions and emerging roles in human cancer. Cellular oncology (Dordrecht). 44: 997-1017.
- Cress WD, Seto E (2000) Histone deacetylases, transcriptional control, and cancer. Journal of cellular physiology 184: 1-16.
- Lian B, Pei YC, Jiang YZ, Xue MZ, Li DQ, et al. (2020) Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics 10: 11092-11109.
- Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, et al. (2010) Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115: 4944-4950.
- Ning Y, Ding J, Sun X, Xie Y, Su M, et al. (2020) HDAC9 deficiency promotes tumor progression by decreasing the CD8(+) dendritic cell infiltration of the tumor microenvironment. Journal for immunotherapy of cancer 8: e000529.
- Ma Z, Liu D, Di S, Zhang Z, Li W (2019) Histone deacetylase 9 downregulation decreases tumor growth and promotes apoptosis in non-small cell lung cancer after melatonin treatment. Journal of pineal research 67: e12587.
- Freese K, Seitz T, Dietrich P, Lee S, Thasler WE, et al. (2019) Histone Deacetylase Expressions in Hepatocellular Carcinoma and Functional Effects of Histone Deacetylase Inhibitors on Liver Cancer Cells In Vitro. Cancers 11: 1587.
- Xu G, Li N, Zhang Y, Zhang J, Xu R, et al. (2019) MicroRNA-3835p inhibits the progression of gastric carcinoma via targeting HDAC9 expression. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas 52: e8341.
- Rastogi B, Kumar A, Raut SK, Panda NK, Rattan V, et al. (2017) Downregulation of miR-377 Promotes Oral Squamous Cell Carcinoma Growth and Migration by Targeting HDAC9. Cancer investigation 35: 152-162.
- Wang W, Liu Z, Zhang X, Liu J, Gui J, et al. (2020) miR-211-5p is down-regulated and a prognostic marker in bladder cancer. The journal of gene medicine 22: e3270.
- Li H, Li X, Lin H, Gong J (2020) High HDAC9 is associated with poor prognosis and promotes malignant progression in pancreatic ductal adenocarcinoma. Molecular medicine 21: 822-832.
- Liang R, Zhang J, Liu Z, Liu Z, Li Q, et al. (2020) Mechanism and Molecular Network of RBM8A-Mediated Regulation of Oxaliplatin Resistance in Hepatocellular Carcinoma. Frontiers in oncology 10: 585452.
© by the Authors & Gavin Publishers. This is an Open Access Journal Article Published Under Attribution-Share Alike CC BY-SA: Creative Commons Attribution-Share Alike 4.0 International License. Read More About Open Access Policy.